FIGURE 4.
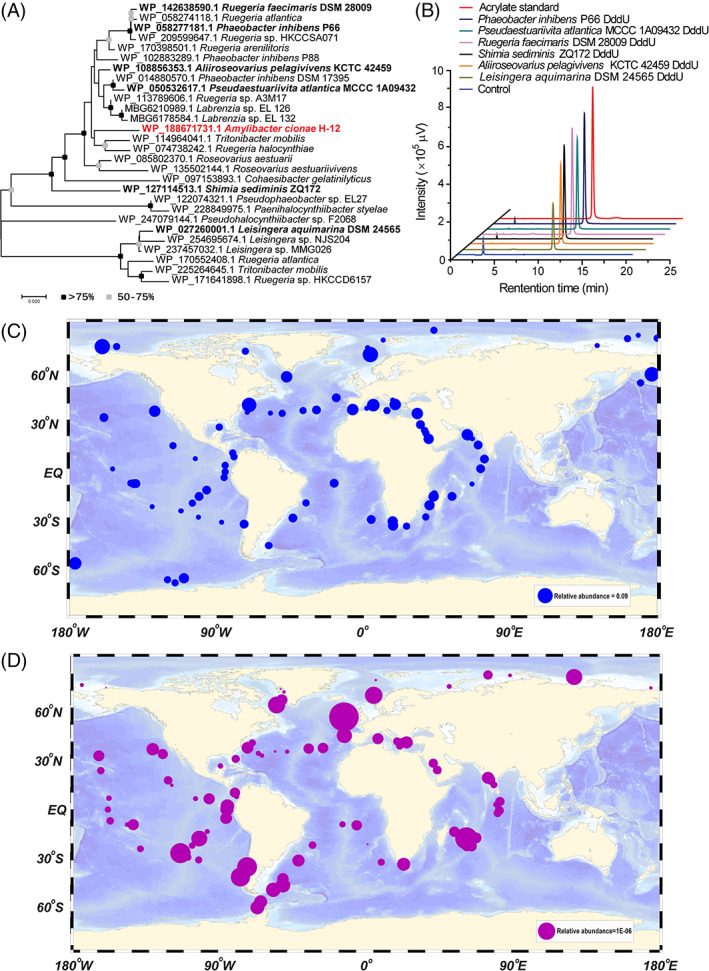
Distribution of DddU in bacteria. (A) The phylogenetic tree of DddU enzymes in Alphaproteobacteria. The black dots represented >75% of the bootstrap values and grey dots represented 50%–75%. Phylogenetic analysis was performed using MEGA version 7.0 (Tamura et al., 2013). Those DddU homologues that are functionally characterized are highlighted in bold. (B) Detection of the dimethylsulfoniopropionate cleavage activity of DddU homologues via high‐performance liquid chromatography. The reaction system without DddU enzymes was used as the control. The acrylate standard was used as a positive control. (C) The geographic distribution of dddU (blue symbols). The relative abundance of dddU at each station was represented by different sizes symbols. (D) The geographic distribution of dddU transcripts (purple symbols). The relative abundance of dddU transcripts at each station was represented by different sizes symbols.
