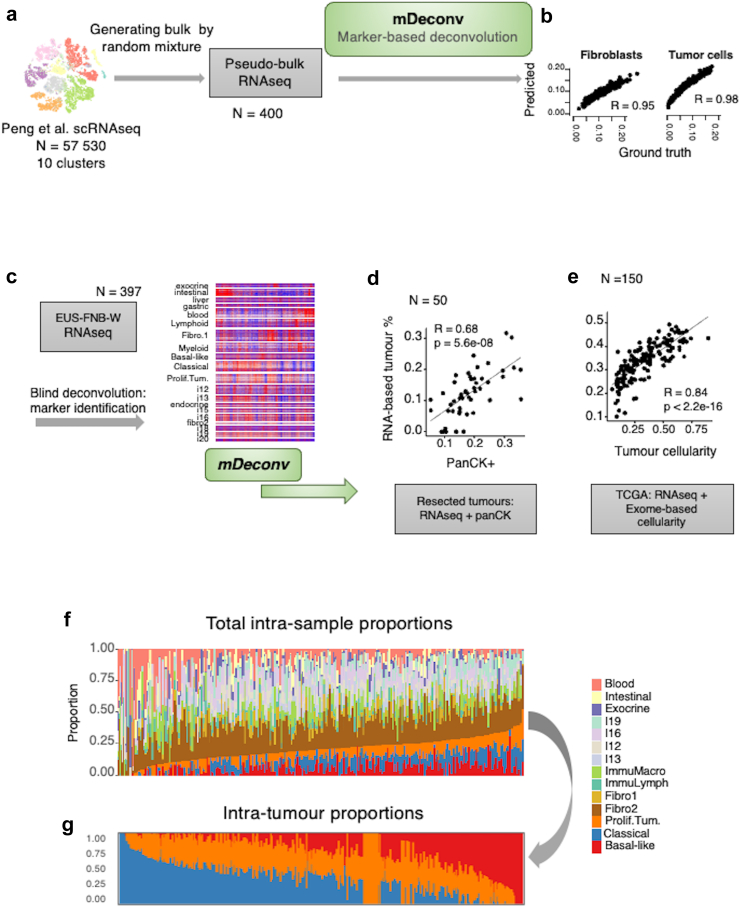
Fig. 3.
RNAseq deconvolution. a: Schematic diagram of the deconvolution method mDeconv on a synthetic dataset. Ten cell types obtained from the clustering of 57,530 single cell RNA-seq were randomly sampled and summed to generate 400 synthetic pseudo-bulk RNA-seq. mDeconv was applied blindly to the synthetic dataset and proportions were estimated from the pseudo-bulk RNA-seq. b: Spearman’s correlations are shown, comparing estimated and true mixture proportions for each of the cell types (fibroblasts and tumour cells). c: Schematic diagram of the application of mDeconv on the BACAP EUS-FNAB-W RNA-seq, identifying 20 independent component-based marker sets. d: Application of mDeconv on a dataset of 50 resected tumours with RNA-seq and corresponding pan-cytokeratin-positive area (panCK+). Scatter plot shows the correlation between the panCK+ and the sum of the estimated proportions of the three RNA-seq-based tumour components. e: Application of mDeconv on the TCGA dataset (150 resected primary tumours). Scatter plot shows the correlation between the exome-based and the RNA-seq estimations of tumour proportions. f: Intra-sample proportions of all the relevant RNA components and g: of tumour components only. The results are from the samples from all the 397 patients.