Figure 2.
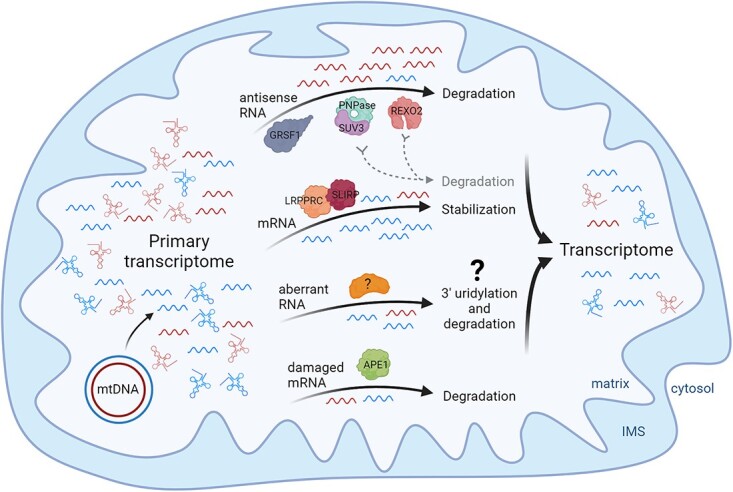
MtRNA decay and surveillance shapes the mitochondrial transcriptome. Pervasive transcription of mtDNA leads to the primary transcriptome which is then shaped by mtRNA degradation. Noncoding antisense mtRNAs are removed in degradosome-dependent manner. The complex is also involved, at least to some extent, in mt-mRNA decay. Mt-mRNAs are bound and stabilized by the LRPPRC/SLIRP complex. Some (abnormal) mitochondrial transcripts undergo uridylation. However, specific role and factors involved in this process are not known. Damaged transcripts with abasic sites are targeted by APE1 and degraded, but the exact mechanism has not been elucidated. The activity of all these decay and surveillance pathways result in formation of the mitochondrial transcriptome. Created with BioRender.com.
