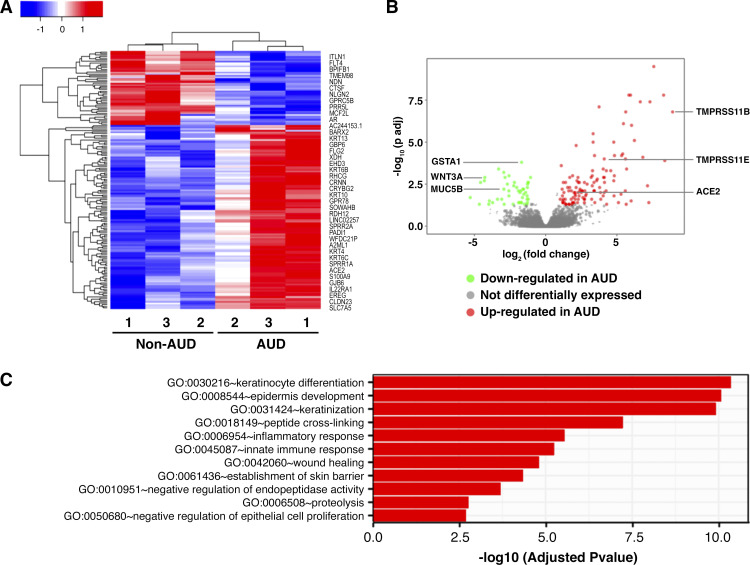
Figure 1.
Differentiated bronchial cells from patients with non-alcohol use disorder (AUD) and AUD were subject to RNA sequencing. A: heat map representation of all 164 differentially expressed genes. Downregulated genes are in blue and upregulated genes are in red, in respect to AUD cells. Select differentially expressed (DE) genes are located on right. B: volcano plot representation of all genes. Green dots represent downregulated genes in AUD cells, red dots represent upregulated genes in AUD cells, and gray dots represent genes that were not differentially expressed. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) receptor genes were upregulated in AUD cells. Genes required for proper function and maintenance of bronchial cells were downregulated in AUD cells. C: differentially expressed genes were clustered by their gene ontology. Enrichment of gene ontology terms was tested using Fisher exact test (GeneSCF v1.1-p2). Shown are top 11 significantly enriched gene ontology terms with an adjusted P value less than 0.05 in the differentially expressed gene set to account for false discovery rate. ACE, angiotensin-converting enzyme.