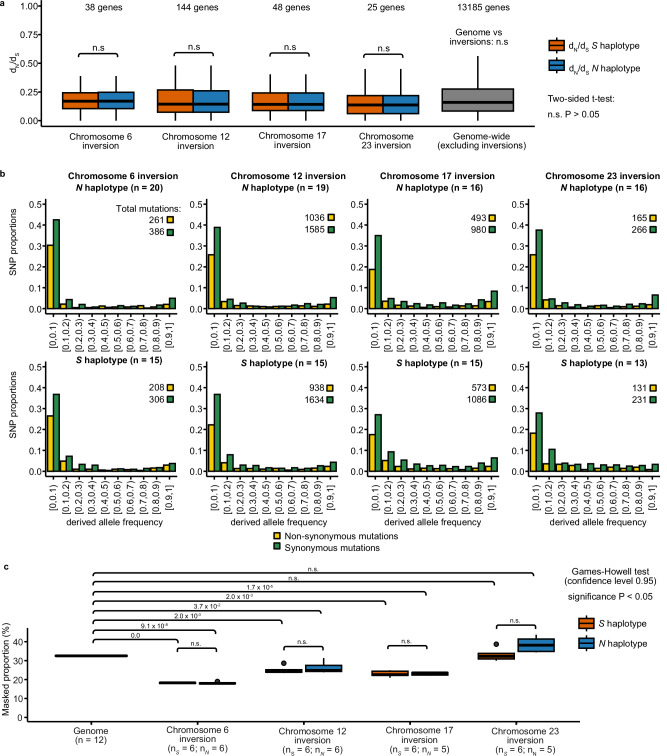
Fig. 7. Lack of mutational load in N and S inversion haplotypes.
a Distribution of dN/dS ratios for N and S alleles of genes overlapping with the inversions on chromosomes 6, 12, 17, and 23 compared with the genome-wide dN/dS distribution. A two-sided t-test revealed no significant differences in the distribution of dN/dS values between N and S alleles, or between inversion haplotypes and the genome. b Site frequency spectra of derived non-synonymous and synonymous mutations for genes inside the inversions, for each inversion haplotype. The total number of mutations in each category is displayed above the graph. c Proportion of transposable elements (TEs) for the entire genome and for each inversion haplotype. A Games-Howell non-parametric two-tailed test was performed to test differences between the proportion of TEs between each allele and the genome (n.s. = non-significant). a, c Boxplots represent the median, 25th and 75th percentiles of distributions.