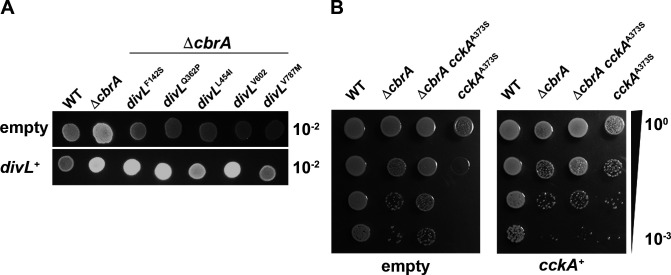
Fig 10.
Complementation assay performed with divL and cckA alleles tests genetic relationship to the wild type. The low copy number pBBR1MCS-3 plasmid (empty) was used to create a complementation plasmid with either divLWT or cckAWT ORF and each with its native promoter region included as predicted by Softberry BPROM. Phenotypes for each strain were accessed using a serial-dilution spot assay on either (A) calcofluor-supplemented LB media grown at 30°C or (B) on LB media grown at 40°C. (A, top row) In the presence of pBBR1MCS-3, the wild type has a calcofluor-dim phenotype, while ΔcbrA is calcofluor-bright. Each ΔcbrA divL double mutant has a calcofluor-dim phenotype relative to ΔcbrA in the presence of pBBR1MCS-3. (A, bottom row) In the presence of pBBR1MCS-3::divLWT, the wild type maintains a calcofluor-dim phenotype, while ΔcbrA remains calcofluor-bright. The divLF142S, divLQ362P, and divLV602M alleles are fully complemented by pBBR1MCS-3::divLWT to the ΔcbrA calcofluor-bright phenotype, which indicates these alleles are recessive. The intermediate calcofluor-bright phenotype of ΔcbrA divLL454I in the presence of pBBR1MCS-3 divLWT indicates this allele is semi-dominate, while the calcofluor-dim phenotype of divLV787M shows that it is dominant. (B, left) During growth at 40°C with pBBR1MCS-3, cckAA373S displays a significant TS growth defect in contrast to ΔcbrA and ΔcbrA cckAA373S mutants. (B, right) During growth at 40°C with pBBR1MCS-3::cckAWT, cckAA373S growth is now similar to other mutant backgrounds, ΔcbrA and ΔcbrA cckAA373S, which indicates this allele has been complemented and is recessive.