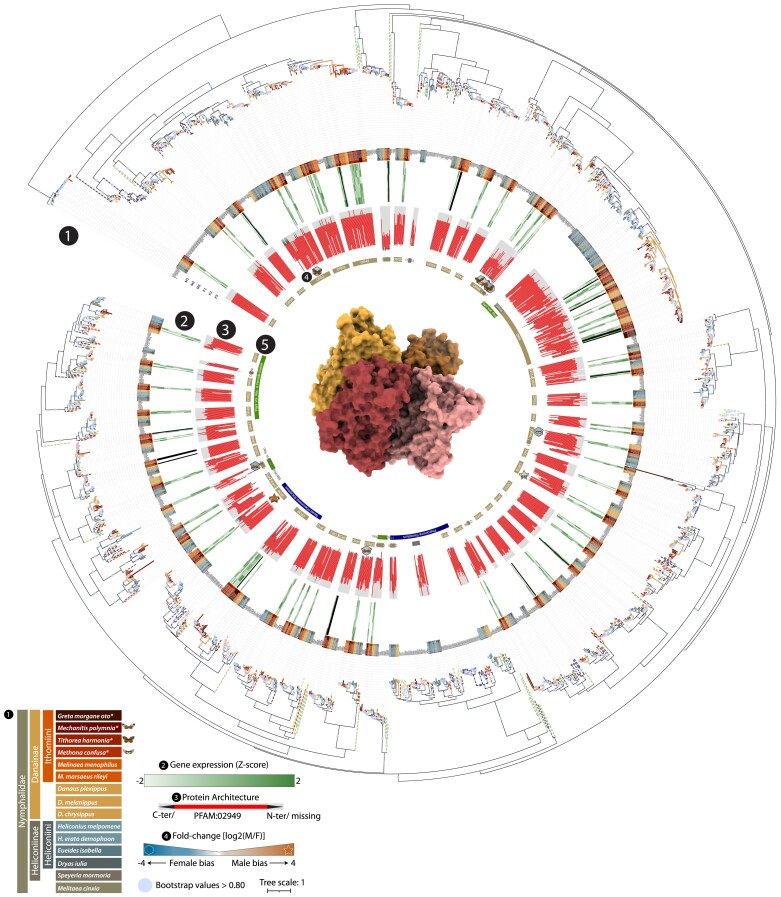
Fig. 3.
Phylogeny of odorant receptor gene family. The ML phylogeny of ORs; the circle size on branches indicates where the bootstrap is higher than 0.80 and are proportional to their values; (1) branch colors are associated to different species, while dashed line to lineages used as references, while sequences annotated in this study are highlighted in the first inner circle. In (2) the heatmap shows the expression levels in the six samples, three males (M1 to 3) and females (F1 to 3); (3) diagram showing the length of the annotated protein and the region that is occupied by the conserved domain. The arrows indicate C-terminus and/or N-terminus that are missing from the protein; (4) fold-change (log2 transformed) of the DEGs, also indicated by the shape (hexagon for female-biased and star for male-biased). The protein structure in the center depicts the general shape of a tetramer of a typical odorant receptor. More detailed information regarding the orthology inference, genomic locations, copy number variations within orthologous groups, and gene expression can be found in supplementary tables S2, S5, and S8, Supplementary Material online. Expression levels can also be observed in Fig. 7a.