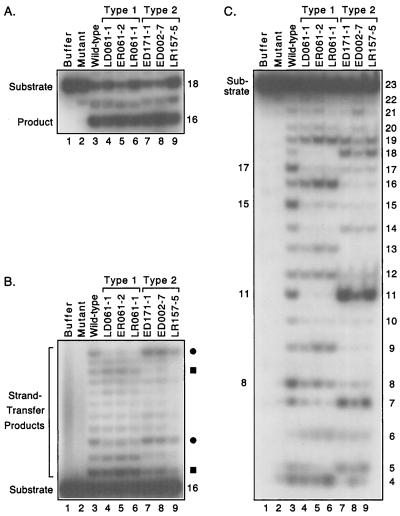
FIG. 2.
Target site selection by patient-derived integrases. Autoradiograms of a denaturing 20% polyacrylamide gel are shown. (A) Processing assay. Double-stranded 18-mers derived from one end of HIV-1 DNA were 5′ labeled on the strand that contains the conserved CA and incubated with protein buffer (lane 1), a laboratory HIV-1 integrase that contained an inactivating D116I mutation (lane 2), the wild-type laboratory HIV-1 integrase (lane 3), or six patient-derived HIV-1 integrases that are grouped as type 1 (lanes 4 to 6) or type 2 (lanes 7 to 9) based on the results shown in panel B. The nomenclature for the naturally occurring integrases is explained in Fig. 3. The 18-mer substrate and the 16-mer product are indicated. The amount of processing in lanes 3 to 9 ranged from 32 to 45%. (B) Joining assay. Double-stranded 16-mers representing a preprocessed end of HIV-1 DNA were 5′ labeled on the strand that contains the conserved CA and incubated with the same proteins as in panel A. The bands indicated by circles or squares are reduced in intensity in the type 1 or type 2 pattern, respectively, compared to the wild-type pattern. The 16-mer substrate and strand-transfer products are indicated. The amount of joining in lanes 3 to 9 ranged from 8 to 23%. (C) Nonspecific alcoholysis assay. Double-stranded 23-mers of nonviral sequence were 5′ labeled on one strand and incubated with the same proteins as in panel A in reactions that included 20% glycerol. Sizes in nucleotides of cleavage products are shown on the right; no bands were detected below the position of 4-mers. The 23-mer substrate and prominent products from the laboratory integrase are indicated on the left. The amount of substrate nicked in lanes 3 to 9 ranged from 41 to 59%.