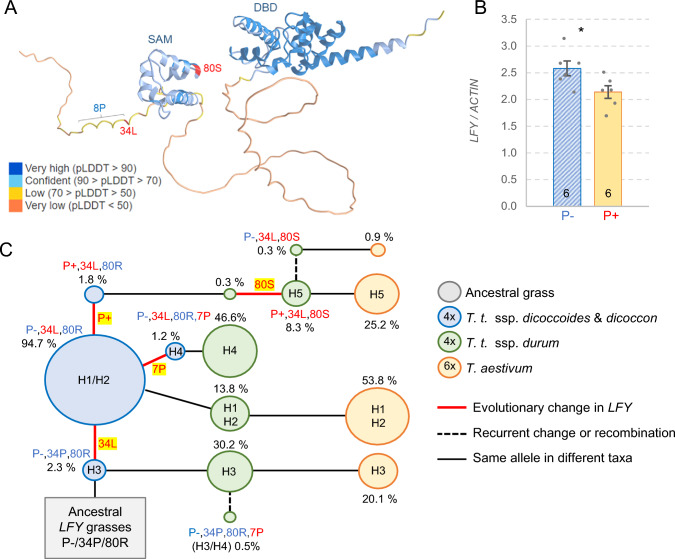
Fig. 2.
Natural variation in LFY-B in wheat. A Location of the natural polymorphisms (in red) in an AlphaFold simulation of the wheat LFY protein for haplotype H5. Colors indicate confidence levels (pLDDT = local distance difference test), SAM = sterile alpha motif oligomerization domain and DBD = DNA binding domain. B quantitative reverse transcription PCR (qRT-PCR) comparison of LFY expression between heterogeneous inbred family (HIF) sister lines homozygous for the H2 (P− 80R, diagonal lines) or H5 (P+ 80S, solid color) haplotypes in developing spikes at the lemma primordia stage. The number of biological replications is indicated at the base of the bars. Error bars indicate standard error of the mean (SEM) and probability values are from two-tailed t-tests. * = P < 0.05. Raw data and statistics are available in Table S7. C Changes in LFY-B allele frequencies in ancestral tetraploid (blue circles), modern tetraploid (green circles), and modern hexaploid (orange circles) wheat accessions. The likely origins of the different polymorphisms are marked in red bold lines and highlighted (color figure online)