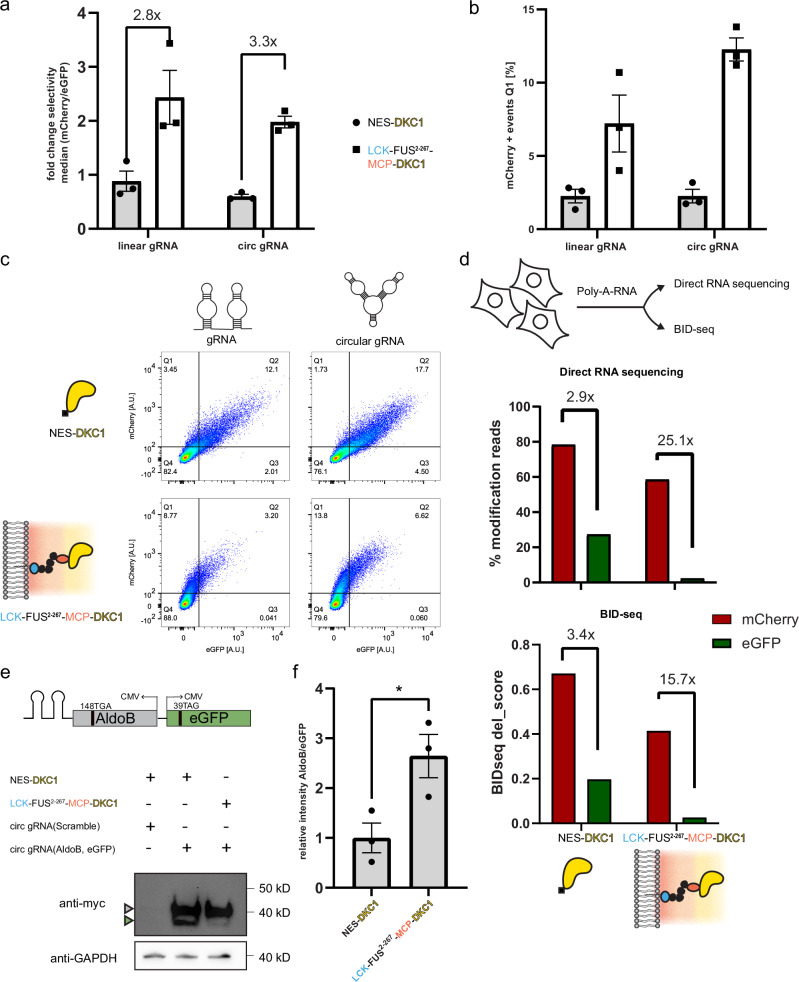
Fig. 4. Circular gRNAs increase designer organelle-mediated pseudouridinylation.
a HEK293T cells were co-transfected with the selectivity reporter (Fig. 1c), gRNAs, and either NES_DKC1 or LCK_FUS2–267_MCP_DKC1.5 × 104 live cells were measured and the fold change in selectivity was calculated for linear and circular gRNAs. b Simultaneously, cells were gated and analyzed for mCherry + cells Q1, as depicted in (c). SEMs of three independent replicates are shown for (a, b). c Representative flow cytometry scatter plots shown for (a, b). d Poly-A RNA was extracted from transfected HEK293T cells and analyzed via direct RNA sequencing using nanopore sequencing via BID-seq. The percentage of modified reads was plotted and the fold change between reporter modifications was compared. Analogous modifications from BID-seq levels (BIDseq del_score: 0–1 refers to unmodified and fully modified, respectively) were compared between NES-DKC1 and OREO transfected cell reporters. e Selective pseudouridinylation of AldoB(148TGA) expressed from a modified selectivity reporter with both proteins tagged with 2× myc tag on the c-terminus. Transfected HEK293T cells were lysed and enriched for full-length AldoB and eGFP via myc-trap beads. A representative western blot of the eluted proteins is shown(top), triangles (gray/AldoB; green/eGFP) indicate the respective protein bands. A small aliquot of lysate was used for myc-trap pull-down and used as a loading control (bottom). Unprocessed blots are available in Source Data file. f Protein bands were quantified and the relative fold change in AldoB/eGFP intensities was calculated. Three independent experiments were performed. An unpaired two-sided t-test was performed (n = 3; p = 0.0359, *p < 0.05) and SEM was plotted. Source data are provided as a Source Data file.