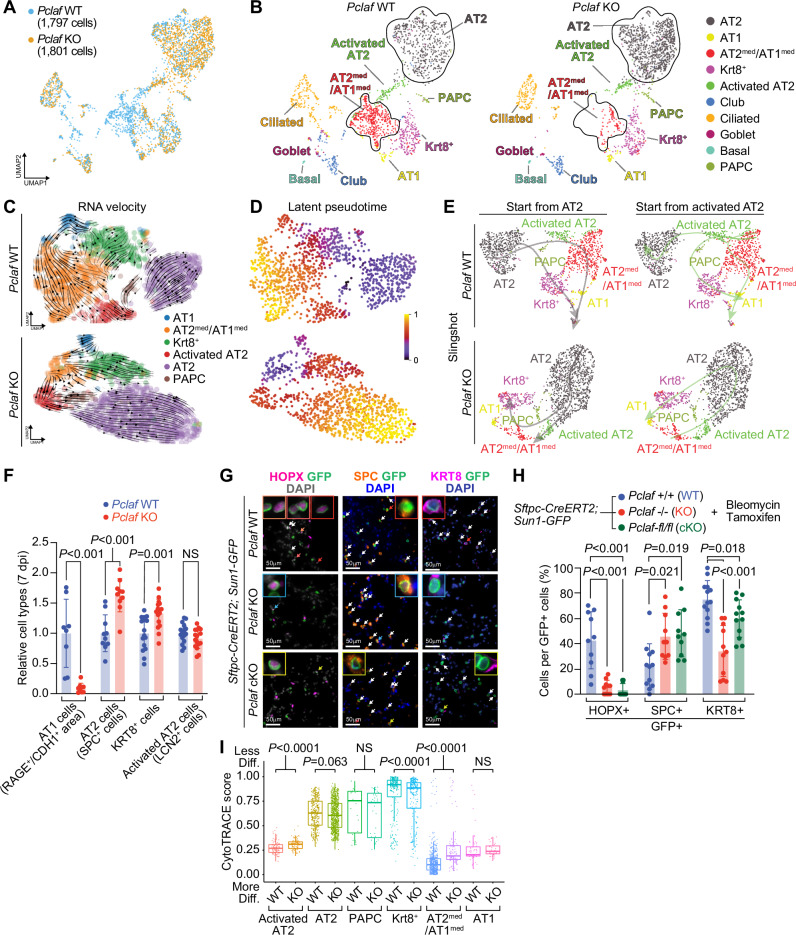
Fig. 3. Pclaf KO suppresses AT2 cell lineage plasticity.
A Integrated UMAP displaying each cell cluster of pulmonary epithelial cells isolated from Pclaf WT and KO mice. B UMAPs (split by Pclaf WT and KO) displaying each cell cluster, colored by cell types. C–E Alveolar cell clusters (AT1, AT2med/AT1med, Krt8+, Activated AT2, AT2, and PAPCs) were introduced for RNA velocity analysis. RNA velocity analyses (C), latent pseudotime analysis (D), and Slingshot analysis (E) of Pclaf WT or KO based on scRNA-seq are depicted. F Quantification of cells derived from AT2 cells immunostaining for RAGE+ area/CDH1+ area (AT1; n = 8 for Pclaf WT and n = 9 for Pclaf KO), SPC+ cells (AT2; n = 10), KRT8+ cells (Krt8+; n = 16), and LCN2+ cells (Activated AT2; n = 16) from the lung tissues (7 dpi). Two-sided Student’s t-test; error bars: mean +/− SD. G, H Quantification of cells derived from AT2 cells using AT2 cell lineage-tracing animal model. The lung tissues of Sftpc-CreERT2; Sun1-GFP lineage-tracing mice combined with Pclaf WT (n = 5), Pclaf KO (n = 3), or Pclaf cKO (Pclaf-fl/fl; n = 3) (14 dpi with tamoxifen for five days) were analyzed by immunostaining (G) and quantification (H). Each cell type was detected by co-immunostaining with an anti-GFP antibody and calculated by their cell numbers per GFP+ cells. HOPX+ (n = 10); SPC+ for Pclaf WT (n = 12), Pclaf KO (n = 10), or Pclaf cKO (n = 10); KRT8+ for Pclaf WT (n = 13), Pclaf KO (n = 11), or Pclaf cKO (n = 12). Sun1-GFP is localized in the inner nuclear membrane. Two-sided Student’s t-test; error bars: mean +/− SD. I Boxplots of predicted cluster-differentiation based on the CytoTRACE analysis using scRNA-seq data shown in (B). The edges of the box plot represent the first and third quartiles, the center line represents the median, and the whiskers extend to the smallest and largest data points within 1.5 interquartile ranges from the edges. Statistical significance is determined using an unpaired Wilcox test. Source data are provided as a Source data file.