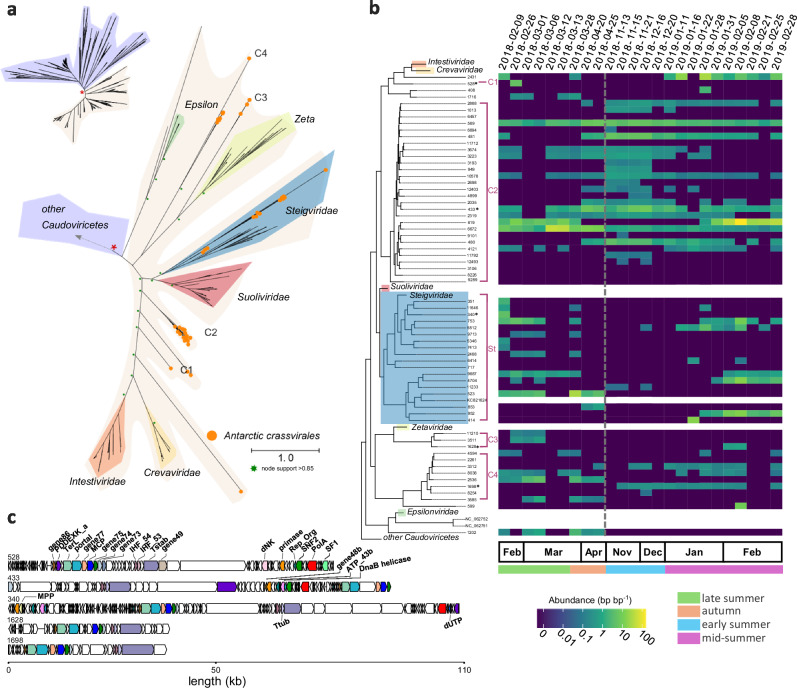
Fig. 4. Diversity and dynamics of Antarctic Crassvirales.
a Unrooted maximum likelihood phylogenetic tree of the terminase large subunit (TerL) of Antarctic Crassvirales scaffolds and reference crassviruses. Crassvirales found in this study are highlighted with the orange circles (n = 62) and shaded area corresponds to the Crassvirales order. b The Crassvirales section of the phylogenetic tree is shown on the left. Reference sequences from the five existing families have been collapsed, except for the Steigviridae that is interspersed with sequences from the Antarctic samples. The vertical coverage (read abundance normalised by scaffold length) for the viral fraction ( < 0.22 µm) are shown as a heatmap.The grey dashed line indicates the winter sampling gap. c Genome map with functional gene annotations for the longest crassviruses of each of the 5 clades, noted in (b) with *. The annotated genes are coloured consistently across the genome maps. Protein abbreviations as follows, major capsid protein (MCP), portal protein, putative structural proteins (genes 73–75, 77 and 86), integration host factor subunits (IHF 53 and 54), tail stabilization protein (Tstab), PD-(D/E)XK family nuclease (PDDEXK), crassvirus uncharacterized gene 49, DnaG family primase, replisome organizer protein (Rep_Org), SNF2 helicase, DNA polymerase family A (PolA), SF1 helicase, AAA domain ATPase (ATP_43b), phage replicative helicase (DnaB), metallophosphatase (MPP), tail tubular protein (Ttub), dUTPase (dUTP).