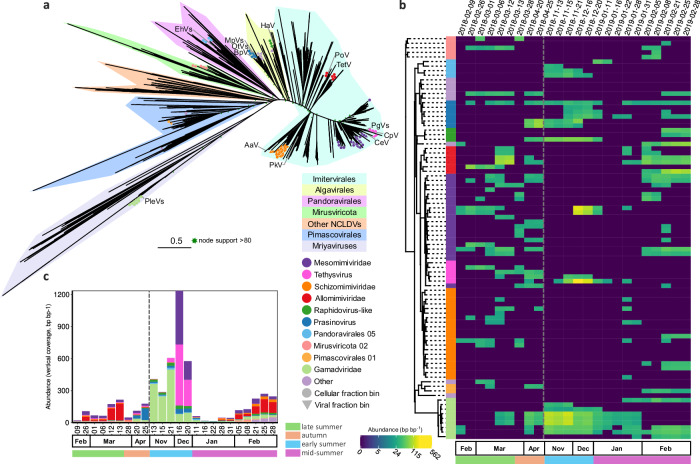
Fig. 5. Diversity and seasonal dynamics of Antarctic NCVs.
a Unrooted maximum likelihood phylogenetic tree of Antarctic NCVs, Mirusviricota and reference genomes from isolates and marine metagenomic surveys based on 7 concatenated marker genes. b Temporal dynamics of sequencing vertical abundance (read abundance normalised by bin length, log-scaled) for the cellular fraction metagenomes (> 0.22 um). The heatmap is organized phylogenetically (rooted between mirusviruses and NCVs) and the first column represents the taxonomy (see panel a). c temporal dynamics at the family level for the cellular fraction metagenomes in bin vertical coverage (read abundance normalised by bin length). The grey dash line indicates the winter sampling gap between April and November 2018. Virus abbreviations as follows, Phaeocystis globosa viruses (PgVs), Chrysochromulina ericina and C. parva viruses (CeV and CpV), Prymnesium kappa virus (PkV), Pyramimonas orientalis virus (PoV) Tetraselmis sp. Virus (TetV), Heterosigma akashiwo virus (HaV), Bathycoccus prasinos (BpV), Ostreococcus tauri (OtV), Micromonas pusilla virus (MpV), Aureococcus anophagefferens virus (AaV) and Emiliania huxleyi viruses (EhVs).