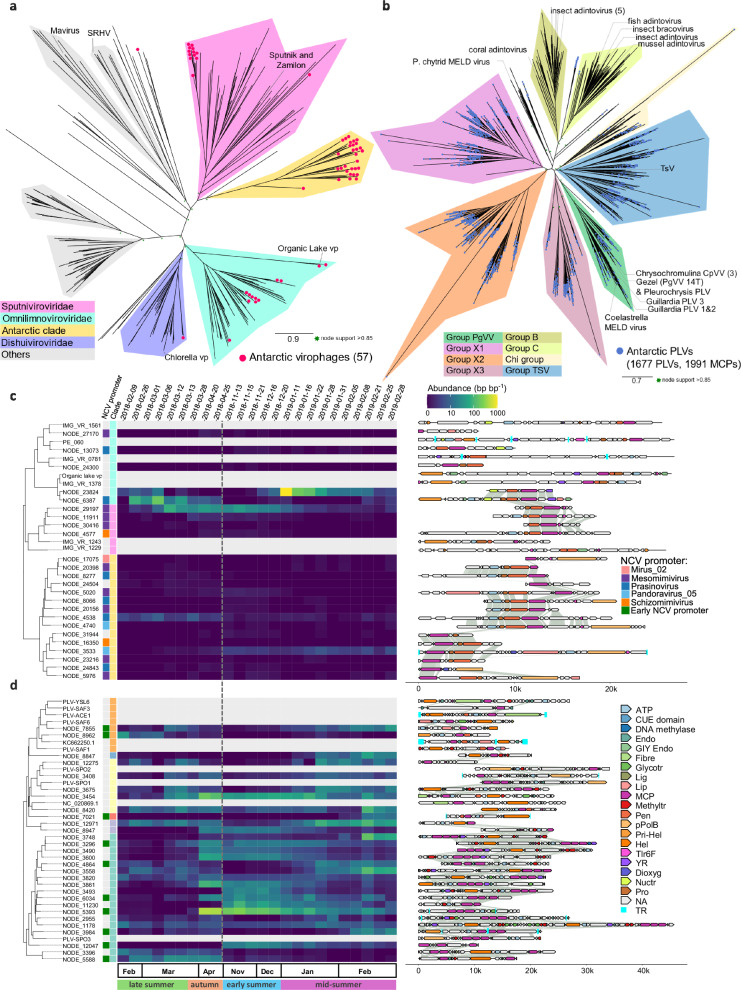
Fig. 6. Antarctic virophages and polinton-like viruses.
Shown are major capsid protein (MCP) phylogenies for (a) virophages (vp, Maverivicetes) and (b) polinton-like viruses (PLV, Polintoviricetes). Virophage family and PLV clade names according to refs. 29,37,38. Phylogeny for (c) virophages longer than 5 kb and (d) top 30 most abundant PLVs longer than 10 kb with the corresponding heatmap of metagenome abundance in the viral fraction (< 0.22 µm) and annotated genome map. Protein abbreviations as follows, FtsK-HerA family DNA-packaging ATPase (ATP), dioxygenase (Dioxyg), endonuclease (Endo), GIY-YIG nuclease domains (GIY), fibre-like protein (Fibre), glycosyltransferase (Glycotr), ligase (Lig), lipase (Lip), methyltransferase (Mthyltr), nucleotidyltransferase (Nuctr), penton (Pen), protein-primed polymerase B (pPolB), primase-helicase (Prim-Hel), helicase (Hel), maturation Cysterine Protease (Pro), uncharacterized protein (Tlr6F), putative tyrosine recombinase (YR). The dash grey line indicates the winter sampling gap.