Fig. 4. Loss of function of Met4 or Met28 affects a different subset of sulphur-responsive genes in C. parapsilosis.
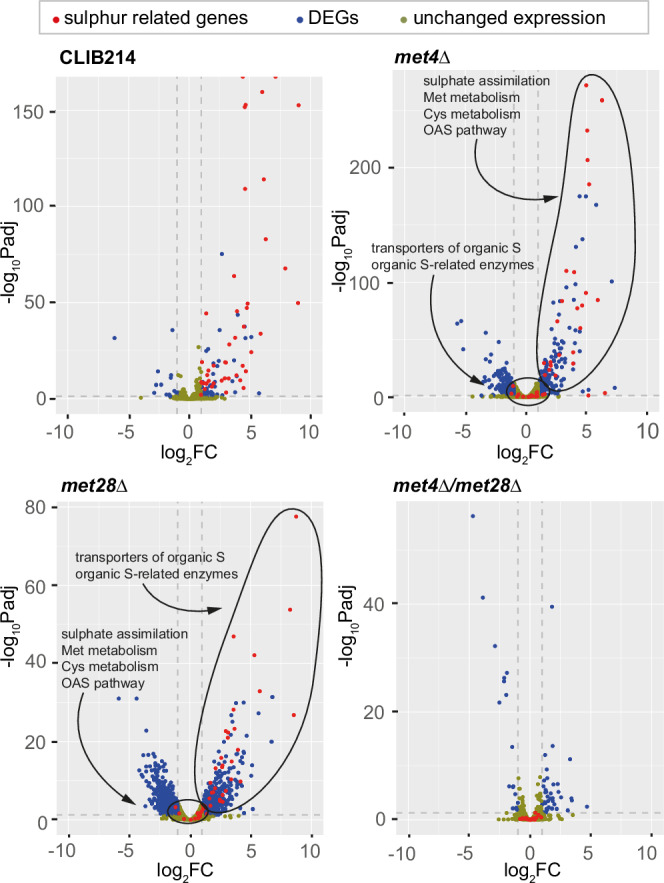
The transcriptional response to cysteine/methionine (C/M) starvation was analysed by RNAseq in C. parapsilosis CLIB214 and strains in which MET4, MET28, or both were deleted. The results are presented in volcano plots (see also Supplementary Data 4, Supplementary Table 2, Supplementary Note 3 and Supplementary Fig. 6). For each gene, the log2 fold change (log2FC) and the -log10 of the adjusted P-value (-log10Padj) are indicated. Values of FC > 2 and adjusted p < 0.05 were selected to define differentially expressed genes (DEGs) between the two conditions (absence of C/M versus presence of C/M). Three dotted grey lines delineate the threshold of significance and fold change. Unaffected genes are coloured light brown, while DEGs are blue. Genes that are implicated in sulphur metabolism are highlighted in red (Supplementary Table 2). Deleting MET4 or MET28 individually affects the expression of two separate subsets of genes, circled in two clusters in the figure (categories as in Supplementary Table 2 and Supplementary Fig. 6), and the absence of both regulators completely shuts down the induction of sulphur-responsive genes: the expression of all the sulphur-related genes collapses below the threshold of significance/fold change in the met4Δ/met28Δ mutant.
