Abstract
The understanding that changes in microbiome composition can influence chronic human diseases and the efficiency of therapies has driven efforts to develop microbiota-centred therapies such as first and next generation probiotics, prebiotics and postbiotics, microbiota editing and faecal microbiota transplantation. Central to microbiome research is understanding how disease impacts microbiome composition and vice versa, yet there is a problematic issue with the term ‘dysbiosis’, which broadly links microbial imbalances to various chronic illnesses without precision or definition. Another significant issue in microbiome discussions is defining ‘healthy individuals’ to ascertain what characterises a healthy microbiome. This involves questioning who represents the healthiest segment of our population—whether it is those free from illnesses, athletes at peak performance, individuals living healthily through regular exercise and good nutrition or even elderly adults or centenarians who have been tested by time and achieved remarkable healthy longevity.
This review advocates for delineating ‘what defines a healthy microbiome?’ by considering a broader range of factors related to human health and environmental influences on the microbiota. A healthy microbiome is undoubtedly linked to gut health. Nevertheless, it is very difficult to pinpoint a universally accepted definition of ‘gut health’ due to the complexities of measuring gut functionality besides the microbiota composition. We must take into account individual variabilities, the influence of diet, lifestyle, host and environmental factors. Moreover, the challenge in distinguishing causation from correlation between gut microbiome and overall health is presented.
The review also highlights the resource-heavy nature of comprehensive gut health assessments, which hinders their practicality and broad application. Finally, we call for continued research and a nuanced approach to better understand the intricate and evolving concept of gut health, emphasising the need for more precise and inclusive definitions and methodologies in studying the microbiome.
Keywords: intestinal microbiology, nutrition, barrier function
Definition of a healthy gut
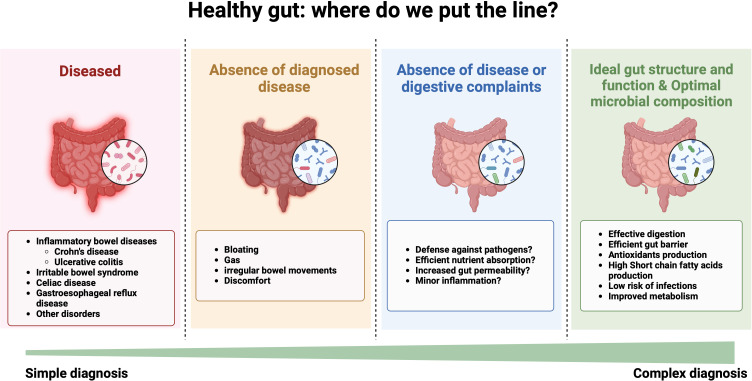
The definition of a healthy gut varies and can be somewhat subjective, as it intertwines with both scientific perspectives and individual health experiences. Almost 15 years ago, the concept of 'gut health' was increasingly used in the medical literature and considered as a major interest in preventive medicine.1 Some experts define a healthy gut from a functional or clinical viewpoint, considering it as not having any diagnosed digestive diseases or disorders. This viewpoint focuses on the lack of detectable medical conditions affecting the gastrointestinal (GI) tract, such as inflammatory bowel diseases (IBD) like Crohn’s disease and ulcerative colitis, irritable bowel syndrome (IBS), coeliac disease, gastroesophageal reflux disease (GERD), and other functional or structural disorders. This definition is pragmatic and clinically useful because it establishes a clear, although basic, criterion for gut health: if an individual does not suffer from any known GI diseases or disorders, their gut could be considered ‘healthy.’ However, this narrow approach may overlook the fact that not all gut health issues manifest in ways that meet the criteria for a formal diagnosis (figure 1). Moreover, it goes beyond the WHO definition of healthy that is more than the mere absence of disease or infirmity and is defined as a state of complete physical, mental and social well-being (https://www.who.int/about/governance/constitution). Importantly, even within this clinical definition, the role of a balanced and diverse gut microbiota in preventing such diseases cannot be overlooked.
Figure 1. Spectrum of gut health from diseased to optimal gut functionality. The definition of a healthy gut varies depending on scientific/medical perspective. The narrowest definition focuses pragmatically on the ‘absence of a diagnosed disease or disorder’. A broader definition, the ‘absence of disease or digestive complaints’, acknowledges subclinical issues. The broadest definition, ‘a healthy gut has an ideal gut structure and function, including an optimal microbial composition’, encompasses the gut’s impact on the host. Created with BioRender.com.
A wider perspective on gut health considers the absence of digestive complaints as a key indicator. Symptoms like bloating, gas, irregular bowel movements and discomfort may not always be linked to a specific disorder but can indicate suboptimal gut health (figure 1). Therefore, a healthy gut, by this definition, is not only ‘free from diagnosed diseases’ but also ‘operates without causing any discomfort or signs of dysfunction’. This definition also implicitly relies on a well-balanced gut microbiota, as imbalances often lead to these discomforts. However, certain indicators of a compromised gut health, like increased permeability of the gut barrier or minor inflammation, might not immediately manifest through clear symptoms (figure 1). Yet, they are still undesirable as they are associated with elevated health risks and/or could lead to the development of diseases or disorders in the future.
The third and most comprehensive interpretation focuses on the ideal structure and function of the gut, encompassing the composition of the gut microbiome, since the microorganisms in the GI tract play a critical role in digestion, nutrient uptake, energy harvest, vitamin synthesis, inflammatory modulation and host immune status. This holistic view covers the physical health and integrity of the GI tract, that should ensure efficient absorption of nutrients and form a robust defence against harmful substances. It also involves the gut’s functional capabilities, like effective digestion and immune response, and a balanced and diverse microbiome (figure 2). This perspective emphasises that microbiome diversity, the presence of beneficial versus harmful bacteria (opportunistic) and the gut’s role in immune function, mental health and disease prevention are crucial for overall well-being. These broader criteria appreciate the gut’s role in overall well-being and recognise that a person might not have a diagnosed GI disorder but still experience suboptimal gut health due to an imbalance in the microbial structure and function, poor dietary habits, stress or other lifestyle factors. This perspective highlights the critical balance and interplay between the gut’s physical structure, its operational functions, and the complex community of microbes that it harbours.
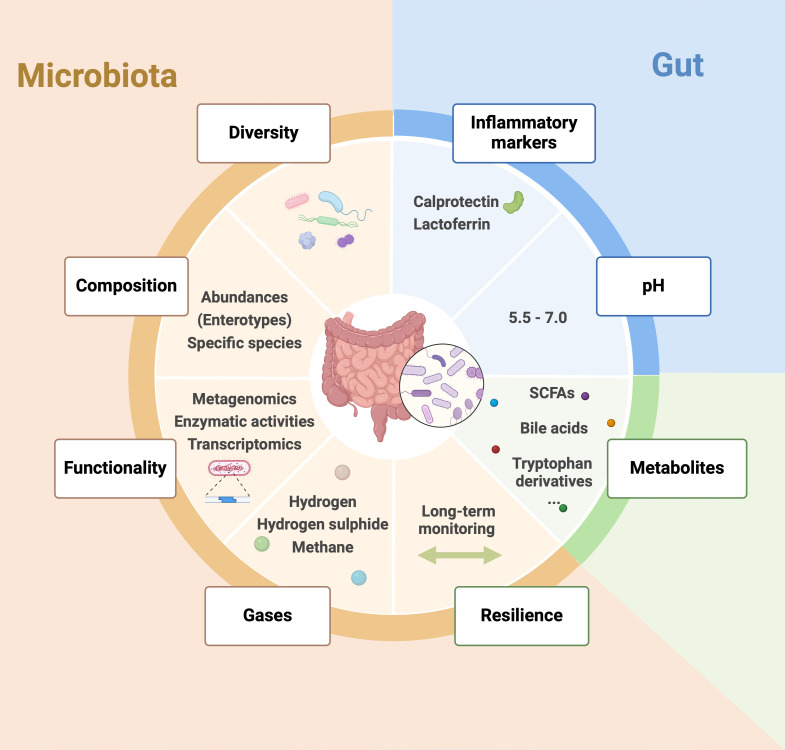
Figure 2. Potential markers of a healthy gut microbiota. A healthy gut microbiota is characterised by a diverse and balanced community of microorganisms that perform vital functions for the host. Several key markers have been proposed to assess these, including microbial diversity, composition (abundances, enterotypes, specific species), functionality (metagenomics, enzymatic activities, transcriptomics) and resilience (through long-term monitoring). Production of important metabolites like SCFAs, bile acids and tryptophan derivatives, as well as gases (hydrogen, hydrogen sulfide, methane), are potential indicators. Additionally, gut health indicators such as pH levels and the presence of inflammation markers (like calprotectin, lactoferrin) can be used for assessing overall gut health. SCFAs, short-chain fatty acids. Created with BioRender.com.
This comprehensive interpretation of gut health contributes to a multifaceted understanding of gut health, emphasising different aspects of what it means to maintain a well-functioning digestive system. However, the framework required for evaluating and achieving optimal GI health is complex and goes beyond mere clinical assessments and everyday experiences of comfort and well-being (figure 1). To fully understand and define a healthy gut microbiome, it is essential to consider these diverse factors and their interdependencies.
The first three lines of defence to maintain host health
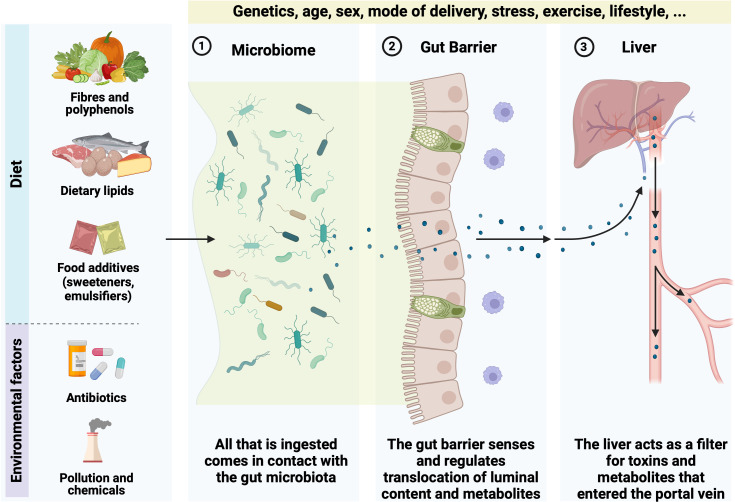
Maintaining health in the face of environmental pressures such as diet, pollutants and toxins is a complex and dynamic process that involves multiple physiological systems (figure 3). Among these, the GI tract and the liver play pivotal roles. The gut microbiota, the gut barrier and the liver represent three critical lines of defence that work synergistically to protect the host from harmful environmental influences.
Figure 3. The three ‘lines of defence’. Diet and environmental factors influence the microbiome (1), which interacts with the gut barrier (2) to regulate the translocation of luminal components and metabolites. The liver (3) acts as a filter for toxins and metabolites entering through the portal vein. An unhealthy state, associated with alterations in gut microbiota composition, may lead to increased intestinal permeability (‘leaky gut’), allowing pathogens and toxins into the bloodstream. Genetics, age, sex and lifestyle factors modulate these processes. Created with BioRender.com.
The gut microbiome
The human microbiome encompasses a diverse and dynamic collection of microbes, including bacteria, fungi and viruses, their genetic material, and by-products that establish residence in our bodies from birth, transferring vertically through generations. The gut microbiota performs several key functions essential for maintaining host health. It aids in the digestion of complex carbohydrates, synthesises essential vitamins and regulates the immune system. The microbiota also competes with pathogenic microbes for resources and space, thereby preventing their colonisation and potential invasion. Moreover, the gut microbiota produces a variety of bioactive compounds, including short-chain fatty acids (SCFAs), which exert anti-inflammatory effects and contribute to the integrity of the gut barrier. The balance and composition of the gut microbiota can be significantly influenced by diet, antibiotics and other environmental factors, making it a crucial mediator in the host’s defence strategy.
Exposure to specific microbes and the interplay with the environment play crucial roles in shaping the microbiome, highlighting the interaction between external factors and our bodies. While colonisation occurs across all body sites, the gut harbours the largest microbial populations.2 In healthy individuals, host factors such as stomach acidity, bile acid production, gut motility and the immune system are classically seen as the main factors influencing the gut microbiome.3,5 However, numerous other variables affect the microbial colonisation, including chemical parameters like intestinal pH, oxygen levels, biological factors like mucus production, antimicrobial molecules and antibody presence6 (figure 2). From birth until approximately the age of 3–4 years, a person develops their primary resident microbiota,7 though the gut microbiome may undergo a more prolonged development, resulting in a composition that is as distinctive as a set of fingerprints.8 Taxonomically, bacteria are classified according to phyla, classes, orders, families, genera and species. The main phyla are Bacillota (formerly Firmicutes), Bacteroidota (formerly Bacteroidetes), Actinobacteriota, Pseudomonadota (formerly Proteobacteria) and Verrucomicrobiota, with the top two phyla Bacillota and Bacteroidota representing roughly 90% of gut microbiota. However, the exact proportion of each of these phyla varies significantly between healthy subjects and do not constitute a major marker of diseases.9 While gaining insight into the composition and dynamics of the human microbiome, particularly within the gut, is crucial for understanding its contribution to health and disease, we suggest emphasising additional critical aspects beyond mere microbiota composition to delineate a healthy gut microbiome.
Over 10 years ago, Lozupone and colleagues highlighted several fundamental questions regarding the diversity of gut microbiota.9 These questions either remained unaddressed or have only been partially explored with the progression of more advanced sequencing technologies. These questions included: What is the extent of diversity within the human microbiota and microbiome (the total gene content within the microbiota) at both the species level and broader taxonomic categories? Among the characteristics of the microbiota, such as specific species or their functions, which are common across many individuals, and which are unique to a single person? And, how effectively can the functions of a microbial community be predicted by understanding the species composition, and we add, and eventually predict the development of human diseases? (figure 2).
The extent of diversity within the human microbiome is vast and varies significantly between individuals. Studies have shown that the human gut microbiome can contain hundreds to thousands of different bacterial species, with significant variation at higher taxonomic levels as well.10 However, despite this diversity, certain core microbial species are present across a large portion of the population, suggesting a common set of functions essential for human health.9
The composition of the gut microbiota generally shows resilience to short-term disturbances, quickly reverting to its original state due to its inherent plasticity.11 12 However, prolonged exposure to stressors common in modern lifestyles, such as Western dietary habits, food additives, environmental contaminants and frequent antibiotic use, can lead to chronic alterations in the gut microbiota. These persistent changes may favour the proliferation of more virulent microorganisms, potentially resulting in negative health outcomes for the host.
There is growing evidence linking alterations in the microbiome to a growing number of diseases, including IBD, liver diseases, obesity, diabetes and even some neurological dysfunctions. The concept of imbalance in the microbiome (as defined below) has been associated with disease states. However, while correlations can be identified, establishing causation and predicting disease development based on microbiome composition are complex tasks.13 14
The composition of gut microbiota is influenced by a complex interplay of both intrinsic (host) and extrinsic (external-environmental) factors, which significantly impact human health. Host-related factors encompass age and delivery mode, liver metabolism capacities, immune system functionality, production of specific bioactive lipids and physiological conditions, where genetic variations contribute to determining microbial diversity and resilience, influencing disease susceptibility and treatment responses. External factors, including dietary habits, hygiene practices, exposure to pollutants and chemicals, psychological stress and lifestyle elements such as physical activity, sleep patterns and social interactions, also play crucial roles in shaping the microbial landscape (figure 3). Diet is a primary determinant and will be discussed more in detail below, though we will not elaborate on the other external influences that have been reviewed elsewhere.
Understanding these complex influences is essential for developing interventions such as personalised nutrition, probiotics, prebiotics and lifestyle modifications to maintain or restore healthy gut microbiota, ultimately enhancing digestive, immune and mental health outcomes. Therefore, we will discuss the role of host factors, including age and delivery, liver metabolism, specific bioactive lipids produced, diet and overall genetics that play significant roles in determining disease risk factors and the resilience of the microbiome.
Characteristics of a healthy gut microbiota
Understanding what constitutes a healthy gut microbiota has been a major focus of recent research. Below we discuss what could be considered as markers of a healthy gut microbiota, their advantages and limitations, and address the ongoing challenges in objectively determining what a healthy gut microbiota should look like.
Diversity
High bacterial diversity, characterised by a large number of different species, is generally considered a marker of good gut health. High microbial diversity contributes to robust digestion, nutrient absorption, metabolite production and immune system regulation. It enhances resilience against disturbances such as antibiotic use and infections and promotes quicker recovery towards a balanced state. Conversely, reduced diversity is linked to various diseases, including IBD, obesity, type 2 diabetes, cardiovascular diseases and certain cancers. Healthy diets and exercise are known to increase microbial diversity, and athletes typically exhibit higher diversity levels.15
However, although low diversity might be indicative of poorer health, high diversity does not necessarily equate to better health. There are indeed notable exceptions to the rule, for example, high gut microbial diversity has been associated with extended colonic transit time and the systemic circulation of potentially harmful protein degradation products.16 Additionally, the beneficial effects of diversity can vary significantly between individuals due to genetic, environmental, and lifestyle differences (figures2 3).
Composition
Rather than merely focusing on diversity, the current literature predominantly examines the specific composition of gut microbiota in diseased versus healthy states. This approach provides deeper insights into the microbial alterations associated with various health conditions, enabling researchers to identify specific bacterial taxa that may contribute to or protect against disease. By comparing the relative abundances and functional roles of these microbial communities, scientists can uncover potential biomarkers for disease and develop targeted therapeutic strategies to modulate the gut microbiome for improved health outcomes.
For example, the Firmicutes/Bacteroidetes (F/B) ratio (now known as Bacillota/Bacteroidota ratio) has garnered significant attention as a potential biomarker for health. Studies have suggested that an increased F/B ratio may be associated with obesity and metabolic disorders, as Firmicutes may be more efficient in energy extraction from food, potentially contributing to weight gain, whereas a higher proportion of Bacteroidetes has been linked to leanness. Additionally, an increased F/B ratio has been observed in GI conditions such as IBD and IBS. However, the F/B ratio’s reliability as a health indicator is limited by individual variability, the microbiome’s complexity and conflicting evidence across studies.17,19
Enterotypes were discovered as robust clusters categorising individuals based on the predominant microbial communities present in their guts.20 This classification was used to provide insights into how different microbial profiles might correlate with various health outcomes, offering a framework for personalised nutrition and medical interventions. Enterotypes have shown to be useful for identifying general patterns in gut health and potential predispositions to certain diseases, thus aiding in the development of targeted therapies and dietary modifications. However, this approach has its limitations and should be used with caution. The concept of enterotypes may oversimplify the complex and dynamic nature of the gut microbiome, which is influenced by numerous factors, including diet, environment and genetics.21,23 Therefore, while enterotypes can provide valuable insights, they should not be the sole metric for assessing gut health, and their use should be complemented with other comprehensive analyses (figure 2).
The relative abundance of certain bacterial genera at lower taxonomic levels could be a more precise indicator of gut health. For instance, a higher prevalence of genera such as Bifidobacterium and Lactobacillus (renamed recently24) is often associated with beneficial gut functions, including improved digestion and enhanced immune response; whereas an over-representation of genera like Clostridium or Escherichia can be indicative of a microbiota deviation and associated with GI disorders, such as IBS or IBD. However, there are significant limitations and counterexamples that challenge the reliability of these indicators. The gut microbiome is highly individualised, influenced by a multitude of factors including diet, genetics and environmental exposures, making it difficult to establish universal biomarkers. Additionally, some genera that are typically considered beneficial can be harmful in specific contexts or in individuals with certain predispositions. For example, an overgrowth of Lactobacillus can contribute to conditions such as small intestinal bacterial overgrowth (SIBO).25 26 Furthermore, the presence of potentially pathogenic genera does not always correlate with disease; some individuals can harbour these bacteria without manifesting any adverse health effects. Therefore, while the relative abundance of bacterial genera provides valuable insights, it must be interpreted with caution, considering the complex and multifaceted nature of the gut microbiome.
Functionality
If compositional diversity alone is insufficient for assessing the health of the microbiota and the host, functional diversity—the range of functions performed by the microbiota—might be more suited to evaluate the continued proper operation of the gut ecosystem. Indeed, a high diversity of species in the gut does not necessarily translate to a wide range of functional capabilities. When comparing composition versus function, studies examining large cohorts have highlighted that while only approximately 45% of bacterial species are similar between two individuals, their microbiota share 82% common metabolic pathways.9 27 28 This significant functional redundancy—capacity of different bacterial species, whether closely related or not, to perform the same functions—suggests that different bacterial compositions can still maintain similar metabolic functions, emphasising the importance of assessing functional diversity over mere compositional diversity for a more comprehensive understanding of gut health. Functional markers such as enzymatic activity and gene content can provide more accurate predictions of physiological states than compositional diversity alone. Enzymatic activity involves the breakdown of complex carbohydrates, proteins, and lipids, while functional gene content reflects the microbiota’s metabolic capabilities. On the other hand, functional redundancy within an individual may itself be an advantage by providing a form of resilience, as the loss of one species may be compensated by another with similar functional capabilities (figure 2).
However, assessing and understanding functional diversity require advanced and expensive techniques such as metagenomics and advanced bioinformatics and often only provide predictive information. Additionally, functional profiles can vary significantly between individuals, complicating the establishment of universal markers.
Metabolites
Products such as SCFAs, bile acids (BAs) and tryptophan metabolites are frequently referred to when assessing gut health and optimal gut microbiota composition. Particularly, SCFAs, including acetate, propionate and butyrate, which are directly produced by gut microbes during the fermentation of dietary fibres, have been extensively studied due to their pivotal roles in maintaining gut barrier integrity, modulating immune responses and serving as an energy source for colonic cells.2 In recent years, a health-promoting role for butyrate has emerged as discussed below. BAs, synthesised from cholesterol in the liver and subsequently modified by gut bacteria, are also valuable indicators of microbial influence on host metabolism and gut integrity. Another significant group is tryptophan metabolites, such as indole and kynurenine, which reflect both microbial and host metabolism, thereby providing a comprehensive picture of gut health.29
Despite their utility, several challenges exist in the accurate quantification of these metabolites, necessitating specialised analytical techniques such as gas chromatography-mass spectrometry and liquid chromatography-mass spectrometry. Additionally, variability in metabolite production due to diet, individual microbiota differences and the complex interplay between microbial and host metabolism can complicate the interpretation of these markers. While SCFAs are reliable indicators of fibre intake and general gut health, their levels can fluctuate based on dietary patterns and individual microbiota composition. Similarly, BAs enterohepatic circulation and host genetic factors can affect their utility as gut health markers. Tryptophan metabolites, although providing broad insights, are influenced by systemic health conditions and dietary intake, adding another layer of complexity.
Overall, while SCFAs, BAs and tryptophan metabolites can certainly offer valuable insights into gut health and microbiota composition, their use as markers is tempered by the need for precise measurement techniques and the influence of various external and internal factors, making it difficult to link specific metabolite levels to distinct microbiota profiles and health states.
Strain specificity
Bacterial strain specificity is crucial due to its significant implications for health, disease and therapeutic interventions. This important concept is often disregarded although under the same microbial species different strains can have vastly different functions. Indeed, the functional diversity among strains mediates differences of efficacy observed in the literature and also partially explains either the deleterious or the beneficial or neutral effects of some bacteria. The best known examples include the case of particular strains of Escherichia coli, like enteropathogenic E. coli (EPEC) and enterohaemorrhagic E. coli (EHEC) that are pathogenic and can cause severe GI disease, whereas other strains are benign and part of the normal gut microbiota.30 Conversely, E. coli Nissle 1917 is considered as a probiotic with anti-inflammatory properties.31 32 Similarly, Helicobacter pylori strains vary in their virulence; strains with the cagA gene are more likely to cause gastric ulcers and cancer.33 Also, Bacteroides fragilis includes strains that produce polysaccharide A (PSA), which modulates the host immune system, while other strains do not. PSA-positive strains can protect against colitis by promoting regulatory T cell function, highlighting the importance of strain-specific interactions in immune modulation.34 Therefore, we urge taking into account the strain specificity when advocating the presence or the lack of effects of some bacteria on a given health situation. It is also for the reason that safety or health claims by regulatory bodies are always related to strains rather than species.
Gases
The production of various gases, such as hydrogen, methane and hydrogen sulfide, is another significant indicator of gut health and microbial composition.29 These gases are by-products of microbial fermentation processes in the gut. Hydrogen and methane are primarily produced by bacterial fermentation of carbohydrates, with methane production being specifically linked to the activity of methanogenic archaea. Hydrogen sulfide, on the other hand, is produced from the metabolism of sulfur-containing amino acids by specific bacteria (figure 2).
The measurement of these gases has been used in clinical settings to diagnose conditions like SIBO and IBS. The rationale for using gas production as a marker is grounded in the fact that specific gas profiles can indicate particular microbial activities and imbalances. For instance, elevated methane levels and methane-producing organisms have been associated with constipation-predominant IBS (IBS-C), while elevated breath levels of hydrogen and hydrogen sulfide as well as a higher relative abundance of hydrogen sulfide-producing bacteria have been found in individuals suffering with diarrhoea-predominant IBS (IBS-D).35
The advantages of using gas production as markers include non-invasive breath tests, which are relatively easy to administer and interpret. However, there are limitations, such as the influence of diet, transit time and individual variability in gas production. Furthermore, interpreting these gases can be complex due to the overlapping symptoms of different GI conditions. Additionally, gases like hydrogen sulfide, despite being indicative of certain bacterial populations, can be difficult to measure accurately due to their reactivity and low concentration in breath samples.
pH levels
In the colon, a pH around 5.5–7 is often associated with a healthy microbiota. An optimal pH environment supports the growth of beneficial bacteria, inhibits pathogenic species, ensures enzymatic functioning, enhances digestive efficiency and nutrient absorption. However, pH levels can fluctuate based on diet, health status and other factors, making it a variable marker. Fermented foods and high-fibre diets promote SCFAs production, beneficially lowering pH in the colon. Certain medications can also influence pH. Therefore, pH alone does not provide a comprehensive picture of gut health and must be considered alongside other markers (figure 2).
Inflammation markers
Low levels of inflammatory markers such as calprotectin and lactoferrin in the stool are indicative of a healthy gut.36 Low inflammation directly reflects the absence of gut inflammation and related disorders. Measuring inflammation markers in stool is a non-invasive method for assessing gut health. However, inflammation markers can be influenced by a wide range of factors, including infections and dietary changes, potentially leading to false positives. Therefore, these markers must be interpreted in the context of other clinical and microbiota data for accurate assessment36 (figures2 3).
Resilience
The ability of the gut microbiota to maintain a stable composition over time and resist disturbances, such as antibiotics or dietary changes, is crucial for gut health. Resilience indicates a robust and adaptable microbiota capable of maintaining homeostasis and supporting overall health. A resilient microbiota can recover quickly from disruptions, reducing the risk of long-term health issues. However, assessing resilience requires long-term monitoring, which can be logistically challenging and costly. Furthermore, the factors contributing to microbiota resilience are complex and not fully understood, complicating the assessment process.
Conclusion
While there are several markers indicating a healthy gut microbiota, there is still no consensus on what constitutes a healthy gut microbiota. Each marker has its advantages and limitations, and the complex interplay between these factors adds to the challenge of defining gut health objectively. Emerging research continues to refine our understanding of these markers and their interactions, but the quest for a definitive and universally applicable definition of a healthy gut microbiota remains ongoing.
The gut barrier
The second line of defence is the gut barrier, a highly selective and dynamic interface between the internal environment and the external world. This barrier consists of a mucus layer, epithelial cells, antimicrobial peptides, tight junctions and immune cells. An intact gut barrier facilitates beneficial interactions between the body and resident gut microbes by enabling efficient nutrient absorption, fending off pathogens and modulating appropriate immune responses. A compromised gut barrier, often resulting from gut microbiota disturbances, inflammation or exposure to toxins, can lead to increased intestinal permeability (referred to as a leaky gut), allowing pathogens and toxins to enter the bloodstream and trigger systemic immune responses (figure 3).
Various bacteria, including Akkermansia muciniphila that lives in the mucus layer, have been found to improve the gut barrier function by increasing the expression of tight junction proteins and hereby reducing the circulating lipopolysaccharides (LPS) endotoxin level. These will be discussed below.
Colonocytes
The colonic epithelial cells, also known as colonocytes, primarily rely on SCFAs like butyrate, which are produced by the fermentation of dietary fibres by gut microbiota. Pioneering studies from Baumler’s team demonstrate that butyrate prompts colonocytes to consume oxygen via the β-oxidation pathway, thereby protecting the host from the proliferation of potentially pathogenic bacteria that can lead to IBD.37 Colonocytes are constantly exposed to numerous microbial antigens and metabolites but generally maintain a symbiotic relationship with these microbes, avoiding inflammation. This symbiosis is particularly notable with obligate anaerobic bacteria that thrive in low-oxygen conditions (hypoxia) and produce butyrate, a crucial energy source for colonocytes, helping to sustain a healthy gut.38 39 Additionally, butyrate reduces immune cell recruitment and proinflammatory signals.40 However, potentially pathogenic bacteria, such as those from the Enterobacteriaceae family, can colonise the gut and cause a microbiota deviation, especially after antibiotic treatment. In that context, the changes in the microbiota composition are often characterised by the bloom of facultative anaerobes, capable of surviving in both oxygen-rich and oxygen-poor environments. These gut microbes are a contributing factor to IBD.41
A healthy gut is characterised by preserving a perfect intestinal equilibrium and thereby promoting the growth of obligate anaerobes over facultative anaerobes. To do so, maintaining hypoxia in the gut lumen is essential to prevent the expansion of facultative anaerobes, such as pathogenic Escherichia and Salmonella.41 Nitrate, produced by the enzyme nitric oxide synthase 2 (NOS2) expressed in colonocytes, serves as an energy source for facultative anaerobes. Therefore, regulating host-derived nitrate and oxygen is crucial for maintaining symbiosis. Recent findings suggest that obligate anaerobes also prevent the expansion of facultative anaerobes by limiting host production of nitrate and oxygen.42 From a mechanistic point of view, this can be achieved when colonocytes are consuming oxygen to β-oxidise butyrate. This contributes to luminal hypoxia by reducing oxygen diffusion into the gut lumen and eventually helps to maintain anaerobic conditions in the lumen. Butyrate also binds to peroxisome proliferator-activated receptor gamma (PPARγ), inhibiting NOS2 and reducing nitric oxide (NO) and nitrate production. Immune cells also respond to butyrate through GPR43 and GPR10940 (for review see Mann et al43). Moreover, PPAR-γ activates mitochondrial β-oxidation in macrophages consuming oxygen as well.44
In pathological conditions, low butyrate levels in the lumen lead to decreased PPARγ activity, increased glycolysis and reduced oxygen consumption. This results in higher NOS2 expression, more NO production and increased nitrate availability for specific pathogens. Recent studies have indicated the importance of butyrate-producing bacteria in the gut. Microbiota analysis of two cohorts showed butyrate producers to be associated with reducing the risk of hospitalisation due to infectious diseases.45 Moreover, this effect may be causal since administration of the butyrate-producer Anaerobutyricum soehngenii was found to improve health in patients with type 2 diabetes .46
Goblet cells and mucus
Among the other components of the gut barrier, it has been shown that the mucus layer plays a major role (for review see Paone and Cani47). Mucus primarily consists of various components, including 90%–95% water, electrolytes, lipids (1%–2%), proteins and other substances. This secretion is dilute, aqueous and viscoelastic due to the presence of specialised proteins known as mucins. Mucins are a family of large, complex, glycosylated proteins. These mucins, which make up 1%–5% of the mucus, are the key structural and functional elements (for review see Paone and Cani46 and Gustafsson and Johansson48). There are different types of mucins in the intestinal tract and besides the composition of the mucins, a healthy gut is characterised by an adequate mucus thickness, which is not (or poorly) penetrable by bacteria. The turnover of the intestinal mucus layer encompasses the processes of mucus synthesis, secretion and degradation. This is a finely tuned mechanism that must be carefully regulated and balanced to maintain proper mucus function (for review see Paone and Cani47 and Gustafsson and Johansson48) (figure 3).
Recent data show that several prebiotics might also contribute to the regulation of the mucus production, composition and degradation. For example, fructo oligosaccharide (FOS) treatment can prevent high-fat diet (HFD)-induced metabolic disorders by stimulating the production of glucagon like peptides 1 and 2 (GLP-1 and GLP-2),49 50 but also likely at least in part, by acting on all the processes of the mucus production.51 Prebiotic treatments like 2′-fucosyllactose (2′FL), an abundant component of human milk oligosaccharides (HMOs), have been shown to enhance gut barrier integrity by influencing the mucus layer and contributes to protection against obesity in vivo.52 In the context of HFD feeding, 2′FL supplementation can counteract obesity and metabolic alterations. It is associated with changes in the intestinal mucus layer, characterised by increased expression of secreted and transmembrane mucins, glycosyltransferases and alterations in mucin glycans composition.52 Therefore, HMOs not only contribute to the maturation of the infant gut barrier,53 but in high concentrations they enhance intestinal epithelial cell function, reduce inflammation and metabolic disorders likely through gut microbiota–mucus barrier dependent mechanisms.52
Immune cells
Maintaining a healthy gut barrier is essential for a healthy gut microbiome, as the gut microbiome plays a pivotal role in our overall health, affecting everything from immune responses to metabolic processes.2 54 Indeed, substantial research has shown that the gut microbiome and the immune system are in constant communication, with gut microbes influencing the activity of our immune defence mechanisms (B cells, T-cells, myeloid cells, innate lymphoid cell (ILC)).55 This interaction is a delicate balance, as imbalances in T-cell activity can lead to a range of health issues, including inflammatory and autoimmune diseases.56 Disruptions in the gut microbiota and the resulting microbiome impact are hallmark traits of IBD. This condition is marked by a notably low presence of butyrate-producing bacteria, including Faecalibacterium spp or Roseburia hominis alongside diminished butyrate levels. Some other bacteria such as Bifidobacterium spp are participating in trophic chains with acetate–lactate converting butyrogens, such as Anaerostipes or Anaerobutyricum spp.
Again, in the context of a healthy gut and to maintain gut immune health, the role of butyrate is crucial (for review see Mann et al43). It regulates the plasticity of type 3 innate lymphoid cells (ILC3) and enhances interleukin-22 (IL-22) production.57 It also plays a significant role in modulating adaptive immunity. Butyrate influences the differentiation of B cells and plasma cells, promoting the production of intestinal IgA. During the differentiation of regulatory T (Treg) cells, butyrate supports the expression of FOXP3 and the secretion of IL-10. Moreover, it shifts the balance between T helper 1 (TH1) cells and TH17 cells by upregulating T-bet expression and downregulating RORγt expression.58 Butyrate and propionate also contribute to the maturation of human monocyte-derived dendritic cells and butyrate stimulates the production of antimicrobial peptides by macrophages.59
The liver
The liver, as the third line of defence, is a vital organ responsible for detoxification and metabolic regulation. It processes and neutralises a wide array of environmental toxins, including pollutants, drugs and metabolic by-products (figure 3). Hepatocytes, the main functional cells of the liver, contain a variety of enzymes, such as cytochrome P450 oxidases, which catalyse the biotransformation of lipophilic compounds into more water-soluble forms that can be excreted via bile or urine. The liver also plays a central role in regulating systemic inflammation and immune responses. Kupffer cells, the liver-resident macrophages, constantly surveil and clear pathogens and debris from the blood. Furthermore, the liver synthesises numerous proteins, including acute-phase reactants and coagulation factors, which are crucial for responding to infections and injuries. The liver’s ability to regenerate and adapt to various stresses is fundamental to its role in maintaining homeostasis and protecting the host from environmental insults.
Liver metabolism and the bidirectional gut–liver axis
The liver and gut microbiota interact closely, highlighting the importance of a bidirectional gut–liver axis in maintaining health and the potential for therapeutic interventions targeting this axis in liver and metabolic disorders.60 The portal vein drains the blood from most parts of the GI tract and even in a healthy setting, it is estimated that approximately 10% of blood metabolites are derived from the gut.28 61 62 Exposure of a healthy liver to (fragments of) commensals has been proposed to contribute to the development and maintenance of liver immunity including various resident leucocyte populations.63 Importantly, BAs production and secretion not only affects immune and metabolic processes in the liver but also crucially modulates the composition and functionality of the gut microbiota thereby contributing to human health.5
Bile acids: prototypic metabolic and gut microbiota-modifying players
The liver produces BAs that are essential for the digestion and absorption of fats and liposoluble vitamins in the intestine. BAs also act as signalling molecules and antimicrobial agents that can influence the composition and function of the gut microbiota.5 64 Changes in BAs composition affect the growth of certain microbial populations over others, thus shaping the gut microbiome. The topic of BAs biology is becoming more and more complex as there might exist hundreds of different BAs and >2000 different bile salt hydrolases supporting the overall importance of these metabolic/immune mediators in physiology, health and disease.5 BAs exert their functions via interference with intracellular nuclear hormone receptors which are ubiquitously expressed in various human tissues. These nuclear receptors include farnesoid X receptor (FXR) and G-protein coupled receptors such as Takeda-G-protein-receptor-5 (TGR5) and are activated by BAs thereby regulating metabolism of glucose and lipids and various innate and adaptive immune processes. Activation of FXR and TGR5 by BAs in the gut affects release of various gut-specific hormones such as peptide YY (PYY) and GLP-1 which are of fundamental importance for regulation of energy and metabolic processes. BAs are conjugated with taurine or glycine within hepatocytes and excreted into the bile duct and when reaching the intestine, especially the colon, metabolised by the gut microbiota towards secondary BAs. There, various intestinal bacteria deconjugate and dehydroxylate the primary BAs cholic acid and chenodeoxycholic acid towards deoxycholic acid and lithocholic acid.65 BAs also affect the structure and function of many intestinal bacteria supporting the growth of certain and inhibiting others. They also regulate the synthesis of intestinal antimicrobial peptides and are crucially involved in the regulation of an intact intestinal barrier.66 The importance of BAs biology is also supported by the fact that the presence of certain BAs in the intestine correlates with murine and human longevity. The gut microbiota of centenarians is enriched in bacteria such as Eubacterium limosum and relatives that protect from trimethylamines and the production of undesired trimethylamine oxide67 68 as well as Odoribacteraceae with the ability to produce unique secondary BAs including various isoforms of lithocholic acid such as iso-, 3-oxo-, allo-, 3-oxoallo- and isoallolithocholic acid.69 Isoallolithocholic acid shows potent antimicrobial effects against Gram-positive (but not Gram-negative) bacteria such as Clostridioides difficile and Enterococcus faecium proposing that BAs pathways might contribute to longevity via such mechanisms.69
BAs as therapeutics have the potential to improve various liver pathologies including metabolic dysfunction-associated steatotic liver disease (MASLD).70 In this recent study,70 the secondary BAs, hyodeoxycholic acid, whose levels are depleted in human MASLD, improved MASLD in various preclinical models and negatively regulated intestinal FXR expression. In the intestine hyodeoxycholic acid modulated the gut microbiome and upregulated certain bacteria such as Parabacteroides distasonis.70 Nie and colleagues recently identified an entirely new microbial BAs, that is, 3-succinylated cholic acid (3-succ CA), which is processed by a β-lactamase derived from B. uniformis. This 3-succ CA is a lumen-restricted metabolite with the potential to increase intestinal A. muciniphila relative levels thereby improving metabolic dysfunction-associated steatohepatitis (MASH) progression, since this symbiont alleviates liver inflammation symptoms in preclinical models.71 72 Importantly humans with progressive MASH exhibited lower intestinal concentrations of 3-succ CA. Interference with nuclear hormone receptors has entered clinical medicine as multiple FXR agonists have been investigated in various liver diseases including immune-mediated disorders such as primary sclerosing cholangitis (PSC), primary biliary cholangitis and MASLD.73 74 Besides BAs many other liver-derived factors such as antimicrobial peptides, gall-bladder-derived surfactant protein D or IgA synthesised in the liver exert their functions partly under the control of commensals and are able to regulate gut microbiota homeostasis in health and disease and thereby contribute to a liver–gut–immune-microbiome axis.75,77
Liver immune function and systemic inflammation
The liver plays a crucial role in immune regulation and therefore can deeply influence the gut microbiome through the modulation of systemic and local immunity.78 For example, the liver can secrete factors that influence gut permeability, inflammation and mucosal immunity, thereby affecting the microbial communities in the gut. The liver also affects the level of systemic inflammation, which in turn can affect the gut microbiome. Chronic liver diseases, such as MASLD are associated with changes in the gut microbiome, potentially due to increased intestinal permeability and the translocation of microbial products into the circulation, which can lead to liver inflammation and alter liver metabolism.79 80
Even in healthy conditions, the liver is continuously exposed to bacterial antigens derived from the intestine despite an intact intestinal gut barrier. Yet, a healthy liver does not produce proinflammatory cytokines such as TNF or IL-1 beta (IL-1β). However, the precursor of one IL-1 family member, that is, IL-1 alpha (IL-1α) is expressed constitutively in a healthy liver and is activated in case of major cell death or acute injury.81 IL-1 family proinflammatory cytokines such as IL-1α or IL-1β belong to the most potent proinflammatory cytokines in an organism and in a healthy liver, which is continuously exposed towards bacterial antigens, IL-1 receptor antagonist (IL-1Ra) is constitutively expressed likely to dampen potentially evolving inflammation.82 83 Also, other mechanisms have been recently proposed which might explain why a healthy liver is protected from inflammation despite continuous antigen exposure.84 In this study, the authors showed that in healthy periportal vein zones, immunosuppressive macrophages produce high levels of the anti-inflammatory cytokine IL-10 and express Marco, a scavenger receptor that restrains proinflammatory pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns. Importantly the presence of this type of macrophages was dependent on the gut microbiota and especially Odoribacteraceae via production of the secondary BAs isoallolithocholic acid, contributed to their presence. The authors furthermore demonstrated that certain liver diseases such as PSC or MASH were associated with a decrease in these immunosuppressive macrophages.84 Exposure towards PAMPs such as the LPS endotoxin which is mainly derived from Gram-negative bacteria remains a continuous threat to a human organism and the liver. As soon as a healthy liver fails to clear gut-derived endotoxins (which appear in the portal vein even in health), it might have consequences for the host as presence of systemic endotoxin results in ‘metabolic endotoxaemia’ causing potentially chronic metabolic inflammation as observed in obesity and related disorders.85 86 Endotoxin exerts its proinflammatory functions via various mechanisms including activation of toll-like receptor 4 (TLR4) and various strategies besides the above discussed immunosuppressive macrophages might be needed to counteract and control endotoxin exposure. Liver expression of LPS-binding protein or soluble CD14 may constitute one local mechanism to neutralise hepatic endotoxin effects.87 It has also been recently demonstrated that intestine-derived high-density cholesterol 3 is able to inactivate endotoxin especially in the portal tract thereby preventing activation of inflammatory cascades in the liver.88 As soon as the liver environment changes, and for example hepatic steatosis develops, various immune mechanisms in the liver might be compromised and endotoxin clearance might be impaired. It has been shown in MASLD that not only liver localisation of endotoxin is increased via co-localisation with TLR4 positive liver macrophages but also endotoxin is detectable in the circulation of those patients.89 90 The liver is also exposed to other PAMPs such as DNA derived from viruses and bacteria. Therefore, this organ plays a fundamental role in protecting a healthy organism from overwhelming inflammation driven by intestinal-derived products including bacteria, viruses and their metabolites. Such a protective function may require various yet undiscovered strategies. In addition, other gut-derived factors including dietary factors such as SCFAs might directly or indirectly affect liver immunity.43
It has been recognised in the past decade that besides endotoxins, commensals and especially bacteria-derived DNA might be present in various extraintestinal tissues such as the liver. Such evidence, however, is largely derived from experimental murine studies. In a recent study, Leinwand and colleagues demonstrated that a liver microbiome is present in healthy mice being enriched by Proteobacteria.63 Interestingly, development of hepatic immunity, especially of natural killer T cells, was directed by certain commensals and included chemokine (C-C motif) ligand 5 (CCL5) signalling. It remains also unclear whether the presence of live bacteria is possible in a healthy human liver, or rather just bacterial fragments such as DNA. Moreover, this all relates to low concentrations of bacterial DNA that may suffer from experimental pitfalls.91Commensals might exert beneficial effects in the liver as shown in germ-free (GF) mice studies as they may maintain liver immune homeostasis and were preventive in certain models of liver fibrosis.92 The role of bacterial components/commensals in extraintestinal tissues, however, remains controversial and needs further studies.93
As soon as the intestinal barrier is disrupted as seen in many chronic disorders, and especially in human obesity and related pathologies, the ‘liver microbiome’ picture might change and indeed a bacterial liver signature has been demonstrated in human MASLD.90 We have recently shown that patients with liver cirrhosis and associated hepatocellular cancer exhibit bacterial DNA not only in the liver but also in the circulation.94 When certain commensals appear in the liver with pathobiont activities they might affect liver health and foster disease.95 Translocation of Enterococcus gallinarum (E. gallinarum) triggers liver autoimmunity and, disease is improved by antibiotics and importantly this pathobiont was also detectable in human autoimmune liver disease.95 Finally, modulation of the gut microbiome might even be beneficial in advanced liver disease even using certain prebiotics such as lactulose.96 This prebiotic not only improved mortality in patients with decompensated liver cirrhosis but also decreased the presence of multidrug-resistant bacteria such as vancomycin-resistant E. faecium.96 The topic of intestinal bacteria and their interaction with the liver in both health and disease is fascinating and might have major implications for a better understanding and management of many liver diseases in the future.
In summary, the liver interaction with the gut microbiome is multifaceted, influencing and being influenced by various metabolic and immune processes. This bidirectional relationship underscores the importance of a healthy gut–liver axis in maintaining overall health and provides insights into potential therapeutic interventions for liver and metabolic disorders.
Factors influencing the gut microbiome and health
Various factors influence the composition and functionality of this microbial ecosystem. These factors include diet, antibiotics and medications, age, genetics, lifestyle factors, geography and environment, infections and diseases, birth methods and early life and exposure to toxins and pollutants (for review see de Vos et al2, Cani et al97 and Korpela and de Vos)98 (figure 3).
Age and mode of delivery
Age and delivery mode play crucial roles in shaping the gut microbiota, which in turn can have profound effects on host health.98 99 Recently, consensus has been reached that the newborn is sterile and there is no evidence for a foetal microbiome.91 From the moment of birth, the gut microbiome begins a dynamic process of development influenced by a variety of factors, including the mode of delivery—vaginal birth or caesarean section—and subsequent environmental exposures such as diet and antibiotics.100 The early gut microbiome of caesarean section delivered infants is unusual and shows reduced relative levels of Bacteroides and Bifidobacterium spp, while so-called pathobionts are increased compared with vaginally born infants.101 102 Since maternal faecal microbiota transplantation but not vaginal seeding of caesarean section born infants can normalise their gut microbiota and reduce the level of pathobionts, it has been proposed that the origin of the newborn’s gut microbiome is the mothers’ gut.103 Recently, the contribution of faecal microbes of the father also has been elucidated and the absence of maternal transmission to the caesarean section delivered newborn is explained by limited physical contact as well as effect of antibiotics.104 The initial colonisation process is notably influenced by whether an infant is breastfed or formula-fed, with breastfed infants typically showing a microbiota dominated by rapidly growing Bifidobacterium spp, which is less diverse but tailored to metabolise human milk oligosaccharides effectively.105 106 During pregnancy, maternal immunity and the metabolites produced by microbes play a crucial role. The transfer of microbes during childbirth, along with the transfer of immune factors, microorganisms and their metabolites through breastfeeding, are essential for the initial microbial exposure and immune system development in early life. Progressive introduction of solid foods induces a diversification of the child’s microbiota that will progressively converge towards its adult composition.107 The arrival or outgrowth of new species will trigger an immune response called 'weaning reaction'. This is necessary for the establishment of an immune system that is, at the same time, protective and tolerant towards commensals.108
Later in life, the gut microbiota shows alterations with ageing that are different between healthy and unhealthy ageing populations. Centenarians, often used as an example of healthy ageing, regardless of their geographical origin, tend to have similar taxonomic and functional modifications of their gut microbiota.109 While useful to gain insight on how the gut microbiota composition is modified in different age settings, these results rarely come from longitudinal studies and should be used with caution. These processes have significant implications for the health of humans, shaping our resistance to diseases and overall health from an early age.110
Diet
Dietary components are among the most significant factors influencing the composition and function of the gut microbiota.111 112 This, in turn, impacts the intestinal health and thereby the gut barrier integrity, inflammation and numerous metabolic processes such as energy balance, glucose and lipids metabolism.2 29 113 114 Given the number of reviews covering this topic, we focused on the main nutrients having shown an impact on gut barrier function and health.
The effects of various dietary components are discussed and include fibres, polyphenols, prebiotics, HMOs, fatty acids, refined sugars, sweeteners, and emulsifiers, on the gut microbiota and associated health outcomes.
Fibres
Dietary fibres, found in fruits, vegetables, whole grains and legumes, are non-digestible carbohydrates that escape the digestion in the upper part of the GI tract and may serve as substrates for microbial fermentation in the colon.115 This fermentation process produces SCFAs, including acetate, propionate and butyrate. These three SCFAs are the most extensively researched microbial metabolites and are crucial in managing host metabolism. They serve as energy sources for intestinal epithelial cells and regulate several physiological functions, such as insulin signalling, lipid metabolism and immune cell differentiation.3754 116,118 Mechanistically, butyrate plays a vital role in controlling the environment of intestinal stem cells and in the renewal process of epithelial cell precursors. SCFAs activate specific G-protein-coupled receptors present on intestinal L-cells, namely GPR41 and GPR43, which in turn stimulate the secretion of GLP-1 and and PYY, leading to appetite-suppressing effects119 (figure 3).
Prebiotics
Among the dietary fibre’s family, prebiotics are playing a major role. Prebiotics are defined as ‘substrates selectively used by host microorganisms that confer a health benefit’.120 Common prebiotics include inulin, FOS, HMOs and galacto-oligosaccharides. These compounds enhance the growth of beneficial bacteria such as Bifidobacteria and Lactobacilli, which, in turn, produce SCFAs and other metabolites beneficial for gut health and systemic functions.121 122 A large body of literature has shown that prebiotics can influence gut health via different mechanisms, including the production of SCFAs. By activating GPR43/41 receptors expressed on the L cells, SCFAs not only enhances the secretion of GLP-1 and PYY as stated above, but also the secretion of GLP-2, a gut peptide known to contribute to maintain the gut barrier function by stimulating the proliferation of intestinal epithelial cells, enhancing tight junctions integrity, stimulating the blood flow.123 Overall, the interaction between gut microbes and these molecular actors helps reduce intestinal permeability, improve insulin secretion and sensitivity, decrease food intake, lower plasma lipids and prevent hepatic steatosis and metabolic endotoxaemia, all of which are associated with reduced inflammation (figure 3).
Polyphenols
Polyphenols are complex bioactive compounds abundant in plant-based foods such as fruits, vegetables, tea, coffee and wine.124 125 The phenolic compounds are categorised into two primary groups, that is the flavonoids and the non-flavonoids. Flavonoids are further divided into anthocyanins, flavanols, flavanones, flavonols and isoflavones. On the other hand, non-flavonoid compounds consist of phenolic acids, stilbenes and lignans (for review see Rodríguez-Daza et al126). Most polyphenols are present in food in the form of esters, glycosides or polymers that cannot be absorbed in their native form, therefore, most polyphenols act locally and are not present in the circulation. Polyphenols are mostly recognised for their antioxidant properties within the gut and their probable role in the prevention of various diseases associated with oxidative stress. However, the genome of certain gut microbes contains a variety of enzymes that play a metabolic role in enhancing the bioavailability and bioactivity of unabsorbed polyphenols. Therefore, also impact on the gut microbiota composition. They promote the growth of beneficial bacteria like Lactiplantibacillus (formerly Lactobacillus, see above), Bifidobacterium, Roseburia and Faecalibacterium spp as well as A. muciniphila but also contribute to inhibiting pathogenic bacteria.126,129 The interaction between polyphenols and gut microbiota also leads to the production of bioactive metabolites that contribute to gut barrier function, anti-inflammatory and anticarcinogenic effects.29 130 131 Phenolic metabolites produced by gut microbes include phenylpropionic acid (PPA), which has shown anti-inflammatory properties and reinforces the gut barrier via aryl hydrocarbon receptor (AhR)-dependent mechanisms.132 133 Hydroxyphenylacetic acid (HPAA) and 4-Hydroxyphenylacetic acid (4-HPAA) are known for their anti-inflammatory and antioxidant properties, potentially also protecting against certain cancers, cardiovascular disease and obesity prevention.132 133 A large variety of other phenolic acid compounds are produced by the gut microbiota from dietary polyphenols, including caffeic acid, ferulic acid and gallic acid that have diverse potential health-protective effects.131 132
Fatty acids
The type of dietary fat consumed significantly influences gut microbiota composition and function. Saturated fatty acids (SFAs), commonly found in animal fats and processed foods, have been associated with a reduction in microbial diversity and an increase in proinflammatory bacteria.85 86 Conversely, omega-3 polyunsaturated fatty acids (PUFAs), found in fish oil and flaxseeds, promote the growth of bacteria such as Bifidobacteria and A. muciniphila, that have been mostly associated with improved health.134 135 PUFAs can be metabolised by bacteria from the Bifidobacteria, Enterobacter, Lactobacillus and Clostridium genera into hydroxy and keto derivatives. Among them, CLA and HYA have demonstrated beneficial effects in murine models of colitis, obesity and cancer by activating PPARy, PPARa, GPR120 and GPR140 and by regulating peristalsis through activation of EP3.136,139
Both dietary cholesterol intake and its plasmatic levels are associated with gut microbiota composition.140 141 Cholesterol is only partially absorbed in the upper intestines and every day, 1–2 g of cholesterol enters the colon, where bacteria that possess cholesterol-degrading enzymes will convert it to coprostanol and, to a lesser extent, coprostanone.142 Although isolating and culturing bacteria that metabolise cholesterol has been challenging, Dysosmobacter and other Oscillibacter genera were recently identified. These bacteria negatively correlate with humans’ stool cholesterol levels and were shown to metabolise cholesterol to coprostanone in vitro.143
Sweeteners
Non-caloric artificial sweeteners (NASs) are frequently used in these products to enhance their taste and stability. Despite being approved by regulatory bodies, some artificial sweeteners pose health risks. Research indicates that saccharin, sucralose and aspartame can cause glucose intolerance more significantly than glucose itself, with saccharin having the most pronounced impact.144 Most NASs pass through the human GI tract without being digested and their encounter with the intestinal microbiota has been linked to changes in the gut microbiota composition and function144 (for review see Gauthier et al145). Metagenomic analysis highlighted an upregulation of pathways involved in LPS biosynthesis, suggesting a potential mechanism by which sweeteners might increase susceptibility to type 2 diabetes.144 The same study also found a positive correlation between sweetener consumption and metabolic indicators such as haemoglobin A1c (HbA1c) and blood glucose levels in humans. High intake of refined sugars and artificial sweeteners can adversely affect gut microbiota composition, leading to gut microbiota deviation,146 reduce microbial diversity and impair glucose metabolism, potentially increasing the risk of metabolic disorders like diabetes146 (for review see Hosseini et al146 and Ruiz-Ojeda et al).147
Emulsifiers
Emulsifiers, commonly used in processed foods to improve texture and shelf life, can negatively impact gut microbiota and gut barrier function. Despite their widespread use, concerns about the safety of emulsifiers have emerged. Specifically, carboxymethylcellulose (CMC) and polysorbate 80 (P80) have been extensively researched for their potential to induce metabolic disorders. Studies have shown that emulsifiers like CMC and P80 disrupt the mucus layer of the gut, leading to increased gut permeability (leaky gut). CMC disrupts the microbial environment by fostering bacterial overgrowth in mice, while P80 facilitates the translocation of E. coli.148 Pioneering research from Gewirtz and Chassaing revealed that exposure to CMC and P80 predisposed mice to low-grade inflammation and metabolic syndrome and increased their susceptibility to colitis in IL10−/− mice through a mechanism involving the alteration of the mucus layer.149 This groundbreaking study demonstrated that specific emulsifiers alter the permeability of the mucus, allowing bacteria to come into closer contact with intestinal epithelial cells. They found that the gut microbiota plays a crucial role, as transferring microbiota from emulsifier-treated mice to GF mice replicated the gut barrier alterations, including changes in the mucus layer, disruption of tight junction proteins and induced metabolic endotoxaemia.149 Moreover, in the absence of microbiota, mice were protected from emulsifier-induced gut barrier dysfunction, low-grade inflammation and subsequent metabolic disorders. These findings were further supported by ex vivo studies using a human gut microbiota simulator (M-SHIME), which showed that CMC and P80 exposure increased the proinflammatory potential by raising bioactive flagellin levels through mechanisms involving the gut microbiota.150 GF mice receiving this altered microbiota exhibited the same metabolic disorders, confirming the role of the gut microbiota in mediating the adverse health effects of emulsifiers. More recently, in mice they found that daily oral administration of A. muciniphila mitigated the phenotypic effects associated with the intake of CMC and P80. These effects included excessive eating, weight gain and dysglycaemia. Furthermore, the administration of A. muciniphila also abolished the low-grade intestinal inflammation induced by CMC and P80 consumption.151 In proof of concept double-blind controlled-feeding study, Chassaing et al investigate in healthy adults, the impact of an emulsifier-free diet compared with the same diet enriched with 15 g of CMC per day for a duration of 11 days.152 Compared with the control group, individuals who consumed CMC experienced a slight increase in abdominal discomfort, a deviation of their gut microbiota composition (ie, reduced diversity) and changes in their faecal metabolome, notably a decrease in SCFAs and free amino acids.152 Finally, based on data from a large prospective cohort of French adults they identified a clear link between the probability of developing type 2 diabetes and the consumption of various food additive emulsifiers commonly present in processed foods.153
Specific bioactive lipids
Bioactive lipids play a significant role in health and diseases, influencing various bodily functions and regulating inflammation, gut barrier integrity, the enteric nervous system, immune responses and metabolic health.154,159
These lipids, including PUFAs, endocannabinoids and related congeners, as well as various families of lipids recently identified, can influence various bodily functions and have been linked to the regulation of inflammation, gut barrier, enteric nervous system, immune response and metabolic health.31 160 161
The gut microbiota interacts with lipids derived from the host endogenous secretions and desquamation, dietary fats and bacterial lipids.162 How host lipids modify the composition and metabolism of gut bacteria is only beginning to be deciphered. Dietary fats impact on the gut microbiota is being investigated and has been discussed in a dedicated paragraph in this review. Genetic modification of the host lipid production also impacts the gut microbiota as shown in models of tissue-specific depletion of NAPE-PLD, a key enzyme to produce endocannabinoids.163 164 As the host-gut microbiota is a bidirectional dialogue, gut microbes reciprocally play an important role in maintaining host lipid homeostasis.
Gut microbiota depletion in GF mice illustrates this importance well. GF mice, despite eating more than conventional mice, present an overall lower body fat content, restored on colonisation, in both males and females.165 The gut microbiota’s impact on host lipid composition is not limited to fat storage as GF mice exhibit a modification of the quantity and/or ratio of various lipids. Triglycerides, phospholipids, sphingolipids, glycerophospholipids, plasmalogens, endocannabinoids, PUFAs and their derivatives were found to be modified in the adipose tissue, plasma, serum, liver, faeces and intestines.136166,171 Moreover, in conventional mice, supplementation with beneficial bacteria like A. muciniphila and Dysosmobacter welbionis has shown improvements of the gut barrier function, reduced metabolic endotoxaemia and body weight, decreased metabolic inflammation and improved glucose tolerance concomitantly to changes in the host bioactive lipids.161 172 173
The gut microbiota mechanisms that influence host lipid homeostasis are still under investigation. These may include modulation of gene expression and epigenetics, and the incorporation of bacterial lipids into host tissues.174,176 The signalling role of bacterial lipids has gained more attraction, mainly with the identification of the numerous effects of SCFAs and secondary BAs.65 174 Recently, the variety of bacterial lipids with bioactive properties was extended to, for example, commendamide, an endocannabinoid-like molecule produced by Bacteroides spp, that has immunomodulatory properties,177 178 N-oeloyl-serinol a GPR119 agonist that stimulates the production of GLP1 and PYY,179 GABA-lipopeptides, produced by E. coli Nissle 1917 and Ligilactobacillus murinus, which are associated with reduced visceral pain in patients with IBS and have analgesic capacities,160 180 and C18-3OH, a PPARy agonist that has anti-inflammatory properties and that is produced by E. coli Nissle 1917 and Holdemanella biformis.31 Membrane lipids, mostly studied for their structural importance, participate in gut microbiota–host communication, particularly in immunology. Specific phospholipids and sphingolipids identified in the membrane of A. muciniphila and Bifidobacterium fragilis have demonstrated immunomodulatory effects in vitro and in vivo.175181,184
As this field is in its early stage, the necessary fat input for a healthy gut microbiota, the optimal level of bacterial lipids beneficial to the host or how widespread their production is across species remain to be determined. The diverse enzymatic capabilities of the gut bacteria allow the production of structurally diverse lipids. Unlike mammalian lipids, bacterial lipids can be odd-chained (frequently 15, 17 or 19 carbons) and display a broader range of (de)saturation and of polar heads groups.181 185 186 This opens the possibilities for new discoveries and a better understanding of the lipid-mediated two-way communication.
In conclusion, the host’s dietary choices profoundly influence the composition and function of the gut microbiota, with significant implications for overall health. Dietary fibres, polyphenols, prebiotics and HMOs generally promote a healthy microbiota and beneficial health outcomes. In contrast, saturated fats, artificial sweeteners and emulsifiers can disrupt microbial balance, impair gut barrier function.
Conclusion and perspectives
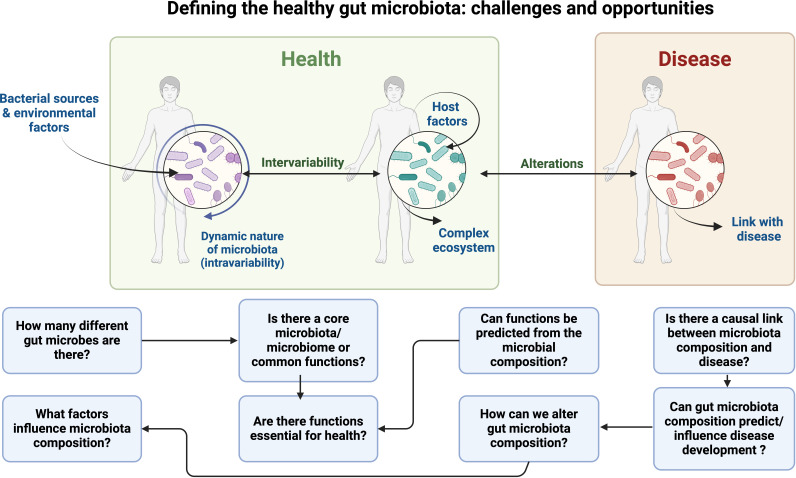
Despite significant advancements in gut microbiome research, defining the healthy gut microbiota remains a formidable challenge (figure 4). The substantial individual variability, influenced by various factor such as genetics, diet, environment and lifestyle, complicates the establishment of a universal standard for a healthy microbiome. Furthermore, the dynamic nature of the gut microbiome, which evolves over time in response to various factors, underscores the difficulty in capturing a single representative snapshot of health.
Figure 4. Challenges in defining a healthy gut microbiota. In health, the gut microbiota is influenced by bacterial sources, environmental and host factors, creating a complex and dynamic ecosystem with intraindividual and interindividual variability. Alterations in this balance can link microbiota to disease. Key research questions address the diversity, influencing factors, essential functions and the potential for microbiota composition to predict or influence disease development. Created with BioRender.com.
The gut microbiome’s immense diversity adds another layer of complexity. Understanding the intricate interactions within this ecosystem and their collective impact on human health is paramount. Current research suggests that the functional capabilities of the microbiome may be more indicative of health than its specific composition, highlighting the need for more precise and affordable methods to identify and measure these functions.
The scarcity of longitudinal data further hinders our understanding, as long-term studies are essential to elucidate the temporal dynamics of the gut microbiome and their implications for health. Additionally, the bidirectional relationship between the gut microbiota and the host’s immune system, metabolism and overall health presents a multifaceted challenge that requires comprehensive investigation.
Environmental and lifestyle factors, such as diet, stress, physical activity and medication use, must be meticulously accounted for when defining a healthy microbiome. These variables, coupled with ethical and practical issues surrounding large-scale data collection and analysis, necessitate standardised methodologies and robust ethical frameworks.
Addressing these challenges will require a multidisciplinary approach, integrating microbiology, genomics, bioinformatics, clinical research and personalised medicine. Future research should focus on developing advanced analytical tools, fostering large-scale longitudinal studies and exploring the functional aspects of the microbiome. Collaborative efforts across scientific disciplines and the incorporation of diverse population data will be crucial in accurately defining and promoting a healthy gut microbiome.
Only by overcoming these challenges can we pave the way for innovative therapeutic strategies and personalised interventions that leverage the gut microbiome to enhance human health and well-being.
Acknowledgements
PDC is honorary research director at FRS-FNRS (Fonds de la Recherche Scientifique) and the recipient of grants from FNRS (Projet de Recherche PDR convention: FNRS T.0030.21, FRFS-WELBIO: WELBIO-CR-2022A-02P, EOS: program no. 40007505). WMdV was supported by the SIAM Gravitation Grant [024.002.002] of the Netherlands Organization for Scientific Research. HT is supported by the excellence initiative VASCage (Centre for Promoting Vascular Health in the Ageing Community), an R&D K-Centre (COMET program - Competence Centers for Excellent Technologies) funded by the Austrian Ministry for Transport, Innovation and Technology, the Austrian Ministry for Digital and Economic Affairs and the federal states Tyrol, Salzburg and Vienna. Research of WMdV was partly supported by the Netherlands Organization for Scientific Research (NWO–SIAM Gravitation Grant 024.002.002) and Business Finland (Grant 329/31/2015). EE-O research is supported by a grant from the Australian Government‘s Medical Research Future Fund, Grant No EPCD000026.
Footnotes
Funding: FNRS (Projet de Recherche PDR-convention: FNRS T.0030.21, FRFS-WELBIO: WELBIO-CR-2022A-02P, EOS: program no. 40007505). SIAM Gravitation Grant [024.002.002] of the Netherlands Organization for Scientific Research. Excellence initiative VASCage (Centre for Promoting Vascular Health in the Ageing Community). R&D K-Centre (COMET program - Competence Centers for Excellent Technologies) funded by the Austrian Ministry for Transport, Innovation and Technology, the Austrian Ministry for Digital and Economic Affairs and the federal states Tyrol, Salzburg and Vienna. The Netherlands Organization for Scientific Research (NWO–SIAM Gravitation Grant 024.002.002) and Business Finland (Grant 329/31/2015). Australian Government’s Medical Research Future Fund, Grant No EPCD000026.
Patient consent for publication: Not applicable.
Ethics approval: Not applicable.
Provenance and peer review: Not commissioned; externally peer reviewed.
Correction notice: This article has been corrected since it published Online First. Author names have been corrected.
Contributor Information
Matthias Van Hul, Email: Matthias.Vanhul@uclouvain.be.
Patrice D Cani, Email: patrice.cani@uclouvain.be.
Camille Petitfils, Email: camille.petitfils@uclouvain.be.
Willem M De Vos, Email: willem.devos@wur.nl.
Herbert Tilg, Email: herbert.tilg@i-med.ac.at.
References
- 1.Bischoff SC. “Gut health”: a new objective in medicine? BMC Med . 2011;9 doi: 10.1186/1741-7015-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Vos WM, Tilg H, Van Hul M, et al. Gut microbiome and health: mechanistic insights. Gut. 2022;71:1020–32. doi: 10.1136/gutjnl-2021-326789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falony G, Joossens M, Vieira-Silva S, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–4. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 4.Procházková N, Falony G, Dragsted LO, et al. Advancing human gut microbiota research by considering gut transit time. Gut. 2023;72:180–91. doi: 10.1136/gutjnl-2022-328166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohanty I, Allaband C, Mannochio-Russo H, et al. The changing metabolic landscape of bile acids – keys to metabolism and immune regulation. Nat Rev Gastroenterol Hepatol . 2024;21:493–516. doi: 10.1038/s41575-024-00914-3. [DOI] [PubMed] [Google Scholar]
- 6.Winter SE, Bäumler AJ. Gut dysbiosis: Ecological causes and causative effects on human disease. Proc Natl Acad Sci U S A. 2023;120:e2316579120. doi: 10.1073/pnas.2316579120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouhy F, Watkins C, Hill CJ, et al. Perinatal factors affect the gut microbiota up to four years after birth. Nat Commun. 2019;10:1517. doi: 10.1038/s41467-019-09252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollister EB, Riehle K, Luna RA, et al. Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome. 2015;3:36. doi: 10.1186/s40168-015-0101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature New Biol. 2012;489:220–30. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leviatan S, Shoer S, Rothschild D, et al. An expanded reference map of the human gut microbiome reveals hundreds of previously unknown species. Nat Commun. 2022;13:3863. doi: 10.1038/s41467-022-31502-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Candela M, Biagi E, Maccaferri S, et al. Intestinal microbiota is a plastic factor responding to environmental changes. Trends Microbiol. 2012;20:385–91. doi: 10.1016/j.tim.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Fassarella M, Blaak EE, Penders J, et al. Gut microbiome stability and resilience: elucidating the response to perturbations in order to modulate gut health. Gut. 2021;70:595–605. doi: 10.1136/gutjnl-2020-321747. [DOI] [PubMed] [Google Scholar]
- 13.Cani PD. Gut microbiota — at the intersection of everything? Nat Rev Gastroenterol Hepatol . 2017;14:321–2. doi: 10.1038/nrgastro.2017.54. [DOI] [PubMed] [Google Scholar]
- 14.Cani PD, Moens de Hase E, Van Hul M. Gut Microbiota and Host Metabolism: From Proof of Concept to Therapeutic Intervention. Microorganisms. 2021;9:1302. doi: 10.3390/microorganisms9061302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohr AE, Jäger R, Carpenter KC, et al. The athletic gut microbiota. J Int Soc Sports Nutr. 2020;17:24. doi: 10.1186/s12970-020-00353-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roager HM, Hansen LBS, Bahl MI, et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat Microbiol. 2016;1:16093. doi: 10.1038/nmicrobiol.2016.93. [DOI] [PubMed] [Google Scholar]
- 17.Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature New Biol. 2006;444:1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 18.Duncan SH, Lobley GE, Holtrop G, et al. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes (Lond) 2008;32:1720–4. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- 19.Khachatryan ZA, Ktsoyan ZA, Manukyan GP, et al. Predominant Role of Host Genetics in Controlling the Composition of Gut Microbiota. PLoS ONE. 2008;3:e3064. doi: 10.1371/journal.pone.0003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature New Biol. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandeputte D, Falony G, Vieira-Silva S, et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016;65:57–62. doi: 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roager HM, Licht TR, Poulsen SK, et al. Microbial enterotypes, inferred by the prevotella-to-bacteroides ratio, remained stable during a 6-month randomized controlled diet intervention with the new nordic diet. Appl Environ Microbiol. 2014;80:1142–9. doi: 10.1128/AEM.03549-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng J, Wittouck S, Salvetti E, et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol. 2020;70:2782–858. doi: 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Zhang R, Ma J, et al. Mucosa-Associated Microbial Profile Is Altered in Small Intestinal Bacterial Overgrowth. Front Microbiol. 2021;12:710940. doi: 10.3389/fmicb.2021.710940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donowitz JR, Parikh HI, Taniuchi M, et al. Increased Fecal Lactobacillus Is Associated With a Positive Glucose Hydrogen Breath Test in Bangladeshi Children. Open Forum Infect Dis. 2019;6:fz266. doi: 10.1093/ofid/ofz266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian L, Wang X-W, Wu A-K, et al. Deciphering functional redundancy in the human microbiome. Nat Commun. 2020;11:6217. doi: 10.1038/s41467-020-19940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Visconti A, Le Roy CI, Rosa F, et al. Interplay between the human gut microbiome and host metabolism. Nat Commun. 2019;10:4505. doi: 10.1038/s41467-019-12476-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Hul M, Neyrinck AM, Everard A, et al. Role of the intestinal microbiota in contributing to weight disorders and associated comorbidities. Clin Microbiol Rev. 2024:e0004523. doi: 10.1128/cmr.00045-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enciso-Martínez Y, González-Aguilar GA, Martínez-Téllez MA, et al. Relevance of tracking the diversity of Escherichia coli pathotypes to reinforce food safety. Int J Food Microbiol. 2022;374:109736. doi: 10.1016/j.ijfoodmicro.2022.109736. [DOI] [PubMed] [Google Scholar]
- 31.Pujo J, Petitfils C, Le Faouder P, et al. Bacteria-derived long chain fatty acid exhibits anti-inflammatory properties in colitis. Gut. 2021;70:1088–97. doi: 10.1136/gutjnl-2020-321173. [DOI] [PubMed] [Google Scholar]
- 32.Hu M, Zhang T, Miao M, et al. Expectations for employing Escherichia coli Nissle 1917 in food science and nutrition. Crit Rev Food Sci Nutr. 2024;2024:1–9. doi: 10.1080/10408398.2023.2301416. [DOI] [PubMed] [Google Scholar]
- 33.Tran SC, Bryant KN, Cover TL. The Helicobacter pylori cag pathogenicity island as a determinant of gastric cancer risk. Gut Microbes. 2024;16:2314201. doi: 10.1080/19490976.2024.2314201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blandford LE, Johnston EL, Sanderson JD, et al. Promoter orientation of the immunomodulatory Bacteroides fragilis capsular polysaccharide A (PSA) is off in individuals with inflammatory bowel disease (IBD) Gut Microbes. 2019;10:569–77. doi: 10.1080/19490976.2018.1560755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villanueva-Millan MJ, Leite G, Wang J, et al. Methanogens and Hydrogen Sulfide Producing Bacteria Guide Distinct Gut Microbe Profiles and Irritable Bowel Syndrome Subtypes. Am J Gastroenterol. 2022;117:2055–66. doi: 10.14309/ajg.0000000000001997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pierre N, Baiwir D, Huynh-Thu VA, et al. Discovery of biomarker candidates associated with the risk of short-term and mid/long-term relapse after infliximab withdrawal in Crohn’s patients: a proteomics-based study. Gut. 2021;70:1450–7. doi: 10.1136/gutjnl-2020-322100. [DOI] [PubMed] [Google Scholar]
- 37.Litvak Y, Byndloss MX, Bäumler AJ. Colonocyte metabolism shapes the gut microbiota. Science. 2018;362:eaat9076. doi: 10.1126/science.aat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roediger WE. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21:793–8. doi: 10.1136/gut.21.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roediger WEW. Review article: nitric oxide from dysbiotic bacterial respiration of nitrate in the pathogenesis and as a target for therapy of ulcerative colitis. Aliment Pharmacol Ther. 2008;27:531–41. doi: 10.1111/j.1365-2036.2008.03612.x. [DOI] [PubMed] [Google Scholar]
- 40.Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature New Biol. 2009;461:1282–6. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Byndloss MX, Olsan EE, Rivera-Chávez F, et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science. 2017;357:570–5. doi: 10.1126/science.aam9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spees AM, Wangdi T, Lopez CA, et al. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. MBio. 2013;4:e00430-13. doi: 10.1128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mann ER, Lam YK, Uhlig HH. Short-chain fatty acids: linking diet, the microbiome and immunity. Nat Rev Immunol. 2024;24:577–95. doi: 10.1038/s41577-024-01014-8. [DOI] [PubMed] [Google Scholar]
- 44.Xavier MN, Winter MG, Spees AM, et al. PPARγ-mediated increase in glucose availability sustains chronic Brucella abortus infection in alternatively activated macrophages. Cell Host Microbe. 2013;14:159–70. doi: 10.1016/j.chom.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kullberg RFJ, Wikki I, Haak BW, et al. Association between butyrate-producing gut bacteria and the risk of infectious disease hospitalisation: results from two observational, population-based microbiome studies. Lancet Microbe. 2024 doi: 10.1016/S2666-5247(24)00079-X. [DOI] [PubMed] [Google Scholar]
- 46.Attaye I, Witjes JJ, Koopen AM, et al. Oral Anaerobutyricum soehngenii augments glycemic control in type 2 diabetes. i Sci. 2024;27:110455. doi: 10.1016/j.isci.2024.110455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paone P, Cani PD. Mucus barrier, mucins and gut microbiota: the expected slimy partners? Gut. 2020;69:2232–43. doi: 10.1136/gutjnl-2020-322260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gustafsson JK, Johansson MEV. The role of goblet cells and mucus in intestinal homeostasis. Nat Rev Gastroenterol Hepatol . 2022;19:785–803. doi: 10.1038/s41575-022-00675-x. [DOI] [PubMed] [Google Scholar]
- 49.Cani PD, Knauf C, Iglesias MA, et al. Improvement of glucose tolerance and hepatic insulin sensitivity by oligofructose requires a functional glucagon-like peptide 1 receptor. Diabetes. 2006;55:1484–90. doi: 10.2337/db05-1360. [DOI] [PubMed] [Google Scholar]
- 50.Cani PD, Possemiers S, Van de Wiele T, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091–103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paone P, Suriano F, Jian C, et al. Prebiotic oligofructose protects against high-fat diet-induced obesity by changing the gut microbiota, intestinal mucus production, glycosylation and secretion. Gut Microbes. 2022;14:2152307. doi: 10.1080/19490976.2022.2152307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paone P, Latousakis D, Terrasi R, et al. Human milk oligosaccharide 2’-fucosyllactose protects against high-fat diet-induced obesity by changing intestinal mucus production, composition and degradation linked to changes in gut microbiota and faecal proteome profiles in mice. Gut. 2024;73:1632–49. doi: 10.1136/gutjnl-2023-330301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matharu D, Ponsero AJ, Lengyel M, et al. Human milk oligosaccharide composition is affected by season and parity and associates with infant gut microbiota in a birth mode dependent manner in a Finnish birth cohort. EBioMedicine. 2024;104:105182. doi: 10.1016/j.ebiom.2024.105182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Régnier M, Van Hul M, Knauf C, et al. Gut microbiome, endocrine control of gut barrier function and metabolic diseases. J Endocrinol. 2021;248:R67–82. doi: 10.1530/JOE-20-0473. [DOI] [PubMed] [Google Scholar]
- 55.Hou K, Wu Z-X, Chen X-Y, et al. Microbiota in health and diseases. Signal Transduct Target Ther. 2022;7:135. doi: 10.1038/s41392-022-00974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ivanov II, Tuganbaev T, Skelly AN, et al. T Cell Responses to the Microbiota. Annu Rev Immunol. 2022;40:559–87. doi: 10.1146/annurev-immunol-101320-011829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang W, Yu T, Huang X, et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Commun. 2020;11:4457. doi: 10.1038/s41467-020-18262-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dupraz L, Magniez A, Rolhion N, et al. Gut microbiota-derived short-chain fatty acids regulate IL-17 production by mouse and human intestinal γδ T cells. Cell Rep. 2021;36:109332. doi: 10.1016/j.celrep.2021.109332. [DOI] [PubMed] [Google Scholar]
- 59.Schulthess J, Pandey S, Capitani M, et al. The Short Chain Fatty Acid Butyrate Imprints an Antimicrobial Program in Macrophages. Immunity. 2019;50:432–45. doi: 10.1016/j.immuni.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tilg H, Adolph TE, Trauner M. Gut-liver axis: Pathophysiological concepts and clinical implications. Cell Metab. 2022;34:1700–18. doi: 10.1016/j.cmet.2022.09.017. [DOI] [PubMed] [Google Scholar]
- 61.Wikoff WR, Anfora AT, Liu J, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guilliams M, Bonnardel J, Haest B, et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell. 2022;185:379–96. doi: 10.1016/j.cell.2021.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leinwand JC, Paul B, Chen R, et al. Intrahepatic microbes govern liver immunity by programming NKT cells. J Clin Invest. 2022;132:e151725. doi: 10.1172/JCI151725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Collins SL, Stine JG, Bisanz JE, et al. Bile acids and the gut microbiota: metabolic interactions and impacts on disease. Nat Rev Microbiol. 2023;21:236–47. doi: 10.1038/s41579-022-00805-x. [DOI] [PubMed] [Google Scholar]
- 65.Fuchs CD, Trauner M. Role of bile acids and their receptors in gastrointestinal and hepatic pathophysiology. Nat Rev Gastroenterol Hepatol. 2022;19:432–50. doi: 10.1038/s41575-021-00566-7. [DOI] [PubMed] [Google Scholar]
- 66.Hofmann AF, Eckmann L. How bile acids confer gut mucosal protection against bacteria. Proc Natl Acad Sci U S A. 2006;103:4333–4. doi: 10.1073/pnas.0600780103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Biagi E, Nylund L, Candela M, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feng Y, Bui TPN, Stams AJM, et al. Comparative genomics and proteomics of Eubacterium maltosivorans: functional identification of trimethylamine methyltransferases and bacterial microcompartments in a human intestinal bacterium with a versatile lifestyle. Environ Microbiol. 2022;24:517–34. doi: 10.1111/1462-2920.15886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sato Y, Atarashi K, Plichta DR, et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature New Biol. 2021;599:458–64. doi: 10.1038/s41586-021-03832-5. [DOI] [PubMed] [Google Scholar]
- 70.Kuang J, Wang J, Li Y, et al. Hyodeoxycholic acid alleviates non-alcoholic fatty liver disease through modulating the gut-liver axis. Cell Metab. 2023;35:1752–66. doi: 10.1016/j.cmet.2023.07.011. [DOI] [PubMed] [Google Scholar]
- 71.Nie Q, Luo X, Wang K, et al. Gut symbionts alleviate MASH through a secondary bile acid biosynthetic pathway. Cell. 2024;187:2717–34. doi: 10.1016/j.cell.2024.03.034. [DOI] [PubMed] [Google Scholar]
- 72.Qu D, Chen M, Zhu H, et al. Akkermansia muciniphila and its outer membrane protein Amuc_1100 prevent high-fat diet-induced nonalcoholic fatty liver disease in mice. Biochem Biophys Res Commun. 2023;684:149131. doi: 10.1016/j.bbrc.2023.149131. [DOI] [PubMed] [Google Scholar]
- 73.Trauner M, Fuchs CD. Novel therapeutic targets for cholestatic and fatty liver disease. Gut. 2022;71:194–209. doi: 10.1136/gutjnl-2021-324305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adorini L, Trauner M. FXR agonists in NASH treatment. J Hepatol. 2023;79:1317–31. doi: 10.1016/j.jhep.2023.07.034. [DOI] [PubMed] [Google Scholar]
- 75.Klag T, Thomas M, Ehmann D, et al. β-Defensin 1 Is Prominent in the Liver and Induced During Cholestasis by Bilirubin and Bile Acids via Farnesoid X Receptor and Constitutive Androstane Receptor. Front Immunol. 2018;9:1735. doi: 10.3389/fimmu.2018.01735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sarashina-Kida H, Negishi H, Nishio J, et al. Gallbladder-derived surfactant protein D regulates gut commensal bacteria for maintaining intestinal homeostasis. Proc Natl Acad Sci U S A. 2017;114:10178–83. doi: 10.1073/pnas.1712837114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moro-Sibilot L, Blanc P, Taillardet M, et al. Mouse and Human Liver Contain Immunoglobulin A-Secreting Cells Originating From Peyer’s Patches and Directed Against Intestinal Antigens. Gastroenterology. 2016;151:311–23. doi: 10.1053/j.gastro.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 78.Anand S, Mande SS. Host-microbiome interactions: Gut-Liver axis and its connection with other organs. NPJ Biofilms Microbiomes. 2022;8:89. doi: 10.1038/s41522-022-00352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang R, Tang R, Li B, et al. Gut microbiome, liver immunology, and liver diseases. Cell Mol Immunol. 2021;18:4–17. doi: 10.1038/s41423-020-00592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Targher G, Byrne CD, Tilg H. MASLD: a systemic metabolic disorder with cardiovascular and malignant complications. Gut. 2024;73:691–702. doi: 10.1136/gutjnl-2023-330595. [DOI] [PubMed] [Google Scholar]
- 81.Chen CJ, Kono H, Golenbock D, et al. Identification of a key pathway required for the sterile inflammatory response triggered by dying cells. Nat Med. 2007;13:851–6. doi: 10.1038/nm1603. [DOI] [PubMed] [Google Scholar]
- 82.Matsukawa A, Fukumoto T, Maeda T, et al. Detection and characterization of IL-1 receptor antagonist in tissues from healthy rabbits: IL-1 receptor antagonist is probably involved in health. Cytokine. 1997;9:307–15. doi: 10.1006/cyto.1996.0170. [DOI] [PubMed] [Google Scholar]
- 83.Tilg H, Adolph TE, Tacke F. Therapeutic modulation of the liver immune microenvironment. Hepatology. 2023;78:1581–601. doi: 10.1097/HEP.0000000000000386. [DOI] [PubMed] [Google Scholar]
- 84.Miyamoto Y, Kikuta J, Matsui T, et al. Periportal macrophages protect against commensal-driven liver inflammation. Nat New Biol. 2024;629:901–9. doi: 10.1038/s41586-024-07372-6. [DOI] [PubMed] [Google Scholar]
- 85.Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–72. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 86.Cani PD, Bibiloni R, Knauf C, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–81. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 87.Wurfel MM, Kunitake ST, Lichenstein H, et al. Lipopolysaccharide (LPS)-binding protein is carried on lipoproteins and acts as a cofactor in the neutralization of LPS. J Exp Med. 1994;180:1025–35. doi: 10.1084/jem.180.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han Y-H, Onufer EJ, Huang L-H, et al. Enterically derived high-density lipoprotein restrains liver injury through the portal vein. Science. 2021;373 doi: 10.1126/science.abe6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Carpino G, Del Ben M, Pastori D, et al. Increased Liver Localization of Lipopolysaccharides in Human and Experimental NAFLD. Hepatology. 2020;72:470–85. doi: 10.1002/hep.31056. [DOI] [PubMed] [Google Scholar]
- 90.Sookoian S, Salatino A, Castaño GO, et al. Intrahepatic bacterial metataxonomic signature in non-alcoholic fatty liver disease. Gut. 2020;69:1483–91. doi: 10.1136/gutjnl-2019-318811. [DOI] [PubMed] [Google Scholar]
- 91.Kennedy KM, de Goffau MC, Perez-Muñoz ME, et al. Questioning the fetal microbiome illustrates pitfalls of low-biomass microbial studies. Nature New Biol. 2023;613:639–49. doi: 10.1038/s41586-022-05546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mazagova M, Wang L, Anfora AT, et al. Commensal microbiota is hepatoprotective and prevents liver fibrosis in mice. FASEB J. 2015;29:1043–55. doi: 10.1096/fj.14-259515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tilg H, Adolph TE. Liver microbes controlling immunity: Facts and pitfalls. Cell Metab. 2022;34:510–2. doi: 10.1016/j.cmet.2022.03.001. [DOI] [PubMed] [Google Scholar]
- 94.Effenberger M, Waschina S, Bronowski C, et al. A gut bacterial signature in blood and liver tissue characterizes cirrhosis and hepatocellular carcinoma. Hepatol Commun . 2023;7:e00182. doi: 10.1097/HC9.0000000000000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Manfredo Vieira S, Hiltensperger M, Kumar V, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. 2018;359:1156–61. doi: 10.1126/science.aar7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Odenwald MA, Lin H, Lehmann C, et al. Bifidobacteria metabolize lactulose to optimize gut metabolites and prevent systemic infection in patients with liver disease. Nat Microbiol. 2023;8:2033–49. doi: 10.1038/s41564-023-01493-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cani PD, Van Hul M, Lefort C, et al. Microbial regulation of organismal energy homeostasis. Nat Metab. 2019;1:34–46. doi: 10.1038/s42255-018-0017-4. [DOI] [PubMed] [Google Scholar]
- 98.Korpela K, de Vos WM. Infant gut microbiota restoration: state of the art. Gut Microbes. 2022;14:2118811. doi: 10.1080/19490976.2022.2118811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Inchingolo F, Inchingolo AD, Palumbo I, et al. The Impact of Cesarean Section Delivery on Intestinal Microbiota: Mechanisms, Consequences, and Perspectives-A Systematic Review. Int J Mol Sci. 2024;25:1055. doi: 10.3390/ijms25021055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jokela R, Korpela K, Jian C, et al. Quantitative insights into effects of intrapartum antibiotics and birth mode on infant gut microbiota in relation to well-being during the first year of life. Gut Microbes. 2022;14:2095775. doi: 10.1080/19490976.2022.2095775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Korpela K, de Vos WM. Early life colonization of the human gut: microbes matter everywhere. Curr Opin Microbiol. 2018;44:70–8. doi: 10.1016/j.mib.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 102.Shao Y, Forster SC, Tsaliki E, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature New Biol. 2019;574:117–21. doi: 10.1038/s41586-019-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Korpela K, Helve O, Kolho K-L, et al. Maternal Fecal Microbiota Transplantation in Cesarean-Born Infants Rapidly Restores Normal Gut Microbial Development: A Proof-of-Concept Study. Cell. 2020;183:324–34. doi: 10.1016/j.cell.2020.08.047. [DOI] [PubMed] [Google Scholar]
- 104.Dubois L, Valles-Colomer M, Ponsero A, et al. Paternal and induced gut microbiota seeding complement mother-to-infant transmission. Cell Host & Microbe. 2024;32:1011–1024.:e4. doi: 10.1016/j.chom.2024.05.004. [DOI] [PubMed] [Google Scholar]
- 105.Lordan C, Roche AK, Delsing D, et al. Linking human milk oligosaccharide metabolism and early life gut microbiota: bifidobacteria and beyond. Microbiol Mol Biol Rev. 2024;88:e0009423. doi: 10.1128/mmbr.00094-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shani G, Hoeflinger JL, Heiss BE, et al. Fucosylated Human Milk Oligosaccharide Foraging within the Species Bifidobacterium pseudocatenulatum Is Driven by Glycosyl Hydrolase Content and Specificity. Appl Environ Microbiol. 2022;88:e0170721. doi: 10.1128/AEM.01707-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roswall J, Olsson LM, Kovatcheva-Datchary P, et al. Developmental trajectory of the healthy human gut microbiota during the first 5 years of life. Cell Host Microbe. 2021;29:765–76. doi: 10.1016/j.chom.2021.02.021. [DOI] [PubMed] [Google Scholar]
- 108.Al Nabhani Z, Eberl G. Imprinting of the immune system by the microbiota early in life. Mucosal Immunol. 2020;13:183–9. doi: 10.1038/s41385-020-0257-y. [DOI] [PubMed] [Google Scholar]
- 109.Ghosh TS, Shanahan F, O’Toole PW. The gut microbiome as a modulator of healthy ageing. Nat Rev Gastroenterol Hepatol . 2022;19:565–84. doi: 10.1038/s41575-022-00605-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Koren O, Konnikova L, Brodin P, et al. The maternal gut microbiome in pregnancy: implications for the developing immune system. Nat Rev Gastroenterol Hepatol . 2024;21:35–45. doi: 10.1038/s41575-023-00864-2. [DOI] [PubMed] [Google Scholar]
- 111.Singh RK, Chang H-W, Yan D, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15:73. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rinninella E, Tohumcu E, Raoul P, et al. The role of diet in shaping human gut microbiota. Best Practice & Research Clinical Gastroenterology . 2023;62–63:101828. doi: 10.1016/j.bpg.2023.101828. [DOI] [PubMed] [Google Scholar]
- 113.Van Hul M, Cani PD. The gut microbiota in obesity and weight management: microbes as friends or foe? Nat Rev Endocrinol. 2023;19:258–71. doi: 10.1038/s41574-022-00794-0. [DOI] [PubMed] [Google Scholar]
- 114.Cani PD, Van Hul M. Gut microbiota in overweight and obesity: crosstalk with adipose tissue. Nat Rev Gastroenterol Hepatol . 2024;21:164–83. doi: 10.1038/s41575-023-00867-z. [DOI] [PubMed] [Google Scholar]
- 115.Cronin P, Joyce SA, O’Toole PW, et al. Dietary Fibre Modulates the Gut Microbiota. Nutrients. 2021;13:1655. doi: 10.3390/nu13051655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vinelli V, Biscotti P, Martini D, et al. Effects of Dietary Fibers on Short-Chain Fatty Acids and Gut Microbiota Composition in Healthy Adults: A Systematic Review. Nutrients. 2022;14:2559. doi: 10.3390/nu14132559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Blaak EE, Canfora EE, Theis S, et al. Short chain fatty acids in human gut and metabolic health. BM. 2020;11:411–55. doi: 10.3920/BM2020.0057. [DOI] [PubMed] [Google Scholar]
- 118.Dalile B, Van Oudenhove L, Vervliet B, et al. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat Rev Gastroenterol Hepatol . 2019;16:461–78. doi: 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- 119.Tolhurst G, Heffron H, Lam YS, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61:364–71. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gibson GR, Hutkins R, Sanders ME, et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol . 2017;14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 121.Delzenne NM, Olivares M, Neyrinck AM, et al. Nutritional interest of dietary fiber and prebiotics in obesity: Lessons from the MyNewGut consortium. Clin Nutr. 2020;39:414–24. doi: 10.1016/j.clnu.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 122.Delzenne NM, Neyrinck AM, Bäckhed F, et al. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat Rev Endocrinol. 2011;7:639–46. doi: 10.1038/nrendo.2011.126. [DOI] [PubMed] [Google Scholar]
- 123.Brubaker PL. Glucagon-like Peptide-2 and the Regulation of Intestinal Growth and Function. Compr Physiol. 2018;8:1185–210. doi: 10.1002/cphy.c170055. [DOI] [PubMed] [Google Scholar]
- 124.Wang X, Qi Y, Zheng H. Dietary Polyphenol, Gut Microbiota, and Health Benefits. Antioxidants (Basel) 11:1212. doi: 10.3390/antiox11061212. n.d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Anhê FF, Choi BSY, Dyck JRB, et al. Host-Microbe Interplay in the Cardiometabolic Benefits of Dietary Polyphenols. Trends Endocrinol Metab. 2019;30:384–95. doi: 10.1016/j.tem.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 126.Rodríguez-Daza MC, Pulido-Mateos EC, Lupien-Meilleur J, et al. Polyphenol-Mediated Gut Microbiota Modulation: Toward Prebiotics and Further. Front Nutr. 2021;8:689456. doi: 10.3389/fnut.2021.689456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Van Hul M, Cani PD. Targeting Carbohydrates and Polyphenols for a Healthy Microbiome and Healthy Weight. Curr Nutr Rep . 2019;8:307–16. doi: 10.1007/s13668-019-00281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pulido-Mateos EC, Lessard-Lord J, Desjardins Y, et al. Biotransformation of camu-camu galloylated ellagitannins by Lactiplantibacillus plantarum with extracellular tannase activity. Food Funct. 2024;15:7189–99. doi: 10.1039/d4fo00149d. [DOI] [PubMed] [Google Scholar]
- 129.Moorthy M, Sundralingam U, Palanisamy UD. Polyphenols as Prebiotics in the Management of High-Fat Diet-Induced Obesity: A Systematic Review of Animal Studies. Foods. 2021;10:299. doi: 10.3390/foods10020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hu J, Chen J, Xu X, et al. Gut microbiota-derived 3-phenylpropionic acid promotes intestinal epithelial barrier function via AhR signaling. Microbiome. 2023;11:102. doi: 10.1186/s40168-023-01551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Man AWC, Zhou Y, Xia N, et al. Involvement of Gut Microbiota, Microbial Metabolites and Interaction with Polyphenol in Host Immunometabolism. Nutrients . 2020;12:3054. doi: 10.3390/nu12103054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kiokias S, Oreopoulou V. A Review of the Health Protective Effects of Phenolic Acids against a Range of Severe Pathologic Conditions (Including Coronavirus-Based Infections) Molecules. 2021;26:5405. doi: 10.3390/molecules26175405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Osborn LJ, Schultz K, Massey W, et al. A gut microbial metabolite of dietary polyphenols reverses obesity-driven hepatic steatosis. Proc Natl Acad Sci USA. 2022;119:e2202934119. doi: 10.1073/pnas.2202934119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Caesar R, Tremaroli V, Kovatcheva-Datchary P, et al. Crosstalk between Gut Microbiota and Dietary Lipids Aggravates WAT Inflammation through TLR Signaling. Cell Metab. 2015;22:658–68. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu F, Smith AD, Solano-Aguilar G, et al. Mechanistic insights into the attenuation of intestinal inflammation and modulation of the gut microbiome by krill oil using in vitro and in vivo models. Microbiome. 2020;8:83. doi: 10.1186/s40168-020-00843-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Druart C, Bindels LB, Schmaltz R, et al. Ability of the gut microbiota to produce PUFA‐derived bacterial metabolites: Proof of concept in germ‐free versus conventionalized mice. Molecular Nutrition Food Res . 2015;59:1603–13. doi: 10.1002/mnfr.201500014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ghosh S, Whitley CS, Haribabu B, et al. Regulation of Intestinal Barrier Function by Microbial Metabolites. Cell Mol Gastroenterol Hepatol. 2021;11:1463–82. doi: 10.1016/j.jcmgh.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Goto T, Kim Y-I, Furuzono T, et al. 10-oxo-12(Z)-octadecenoic acid, a linoleic acid metabolite produced by gut lactic acid bacteria, potently activates PPARγ and stimulates adipogenesis. Biochem Biophys Res Commun. 2015;459:597–603. doi: 10.1016/j.bbrc.2015.02.154. [DOI] [PubMed] [Google Scholar]
- 139.Miyamoto J, Mizukure T, Park S-B, et al. A Gut Microbial Metabolite of Linoleic Acid, 10-Hydroxy-cis-12-octadecenoic Acid, Ameliorates Intestinal Epithelial Barrier Impairment Partially via GPR40-MEK-ERK Pathway. J Biol Chem. 2015;290:2902–18. doi: 10.1074/jbc.M114.610733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature New Biol. 2018;555:210–5. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 141.Le Roy T, Lécuyer E, Chassaing B, et al. The intestinal microbiota regulates host cholesterol homeostasis. BMC Biol. 2019;17:94. doi: 10.1186/s12915-019-0715-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kenny DJ, Plichta DR, Shungin D, et al. Cholesterol Metabolism by Uncultured Human Gut Bacteria Influences Host Cholesterol Level. Cell Host Microbe. 2020;28:245–57. doi: 10.1016/j.chom.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li C, Stražar M, Mohamed AMT, et al. Gut microbiome and metabolome profiling in Framingham heart study reveals cholesterol-metabolizing bacteria. Cell. 2024;187:1834–52. doi: 10.1016/j.cell.2024.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Suez J, Korem T, Zeevi D, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature New Biol. 2014;514:181–6. doi: 10.1038/nature13793. [DOI] [PubMed] [Google Scholar]
- 145.Gauthier E, Milagro FI, Navas-Carretero S. Effect of low-and non-calorie sweeteners on the gut microbiota: A review of clinical trials and cross-sectional studies. Nutrition. 2024;117:112237. doi: 10.1016/j.nut.2023.112237. [DOI] [PubMed] [Google Scholar]
- 146.Hosseini A, Barlow GM, Leite G, et al. Consuming artificial sweeteners may alter the structure and function of duodenal microbial communities. i Sci. 2023;26:108530. doi: 10.1016/j.isci.2023.108530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ruiz-Ojeda FJ, Plaza-Díaz J, Sáez-Lara MJ, et al. Effects of Sweeteners on the Gut Microbiota: A Review of Experimental Studies and Clinical Trials. Adv Nutr. 2019;10:S31–48. doi: 10.1093/advances/nmy037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Roberts CL, Keita AV, Duncan SH, et al. Translocation of Crohn’s disease Escherichia coli across M-cells: contrasting effects of soluble plant fibres and emulsifiers. Gut. 2010;59:1331–9. doi: 10.1136/gut.2009.195370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chassaing B, Koren O, Goodrich JK, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature New Biol. 2015;519:92–6. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chassaing B, Van de Wiele T, De Bodt J, et al. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut. 2017;66:1414–27. doi: 10.1136/gutjnl-2016-313099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Daniel N, Gewirtz AT, Chassaing B. Akkermansia muciniphila counteracts the deleterious effects of dietary emulsifiers on microbiota and host metabolism. Gut. 2023;72:906–17. doi: 10.1136/gutjnl-2021-326835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chassaing B, Compher C, Bonhomme B, et al. Randomized Controlled-Feeding Study of Dietary Emulsifier Carboxymethylcellulose Reveals Detrimental Impacts on the Gut Microbiota and Metabolome. Gastroenterology. 2022;162:743–56. doi: 10.1053/j.gastro.2021.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Salame C, Javaux G, Sellem L, et al. Food additive emulsifiers and the risk of type 2 diabetes: analysis of data from the NutriNet-Santé prospective cohort study. The Lancet Diabetes & Endocrinology. 2024;12:339–49. doi: 10.1016/S2213-8587(24)00086-X. [DOI] [PubMed] [Google Scholar]
- 154.Michalak A, Mosińska P, Fichna J. Polyunsaturated Fatty Acids and Their Derivatives: Therapeutic Value for Inflammatory, Functional Gastrointestinal Disorders, and Colorectal Cancer. Front Pharmacol. 2016;7:459. doi: 10.3389/fphar.2016.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chiurchiù V, Leuti A, Maccarrone M. Bioactive Lipids and Chronic Inflammation: Managing the Fire Within. Front Immunol. 2018;9:38. doi: 10.3389/fimmu.2018.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Abot A, Wemelle E, Laurens C, et al. Identification of new enterosynes using prebiotics: roles of bioactive lipids and mu-opioid receptor signalling in humans and mice. Gut. 2021;70:1078–87. doi: 10.1136/gutjnl-2019-320230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cho YK, Lee S, Lee J, et al. Lipid remodeling of adipose tissue in metabolic health and disease. Exp Mol Med . 2023;55:1955–73. doi: 10.1038/s12276-023-01071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Garcia C, Andersen CJ, Blesso CN. The Role of Lipids in the Regulation of Immune Responses. Nutrients. 2023;15:3899. doi: 10.3390/nu15183899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mantel M, Derkinderen P, Bach-Ngohou K, et al. Crosstalk between omega-6 oxylipins and the enteric nervous system: Implications for gut disorders? Front Med. 2023;10:1083351. doi: 10.3389/fmed.2023.1083351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Petitfils C, Maurel S, Payros G, et al. Identification of bacterial lipopeptides as key players in IBS. Gut. 2023;72:939–50. doi: 10.1136/gutjnl-2022-328084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Moens de Hase E, Petitfils C, Alhouayek M, et al. Dysosmobacter welbionis effects on glucose, lipid, and energy metabolism are associated with specific bioactive lipids. J Lipid Res. 2023;64:100437. doi: 10.1016/j.jlr.2023.100437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gérard P. The crosstalk between the gut microbiota and lipids. OCL . 2020;27:70. doi: 10.1051/ocl/2020070. [DOI] [Google Scholar]
- 163.Everard A, Plovier H, Rastelli M, et al. Intestinal epithelial N-acylphosphatidylethanolamine phospholipase D links dietary fat to metabolic adaptations in obesity and steatosis. Nat Commun. 2019;10:457. doi: 10.1038/s41467-018-08051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Geurts L, Everard A, Van Hul M, et al. Adipose tissue NAPE-PLD controls fat mass development by altering the browning process and gut microbiota. Nat Commun. 2015;6:6495. doi: 10.1038/ncomms7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bäckhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–23. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Brown EM, Clardy J, Xavier RJ. Gut microbiome lipid metabolism and its impact on host physiology. Cell Host & Microbe . 2023;31:173–86. doi: 10.1016/j.chom.2023.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Manca C, Boubertakh B, Leblanc N, et al. Germ-free mice exhibit profound gut microbiota-dependent alterations of intestinal endocannabinoidome signaling. J Lipid Res. 2020;61:70–85. doi: 10.1194/jlr.RA119000424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kindt A, Liebisch G, Clavel T, et al. The gut microbiota promotes hepatic fatty acid desaturation and elongation in mice. Nat Commun. 2018;9:3760. doi: 10.1038/s41467-018-05767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Velagapudi VR, Hezaveh R, Reigstad CS, et al. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res. 2010;51:1101–12. doi: 10.1194/jlr.M002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Liebisch G, Plagge J, Höring M, et al. The effect of gut microbiota on the intestinal lipidome of mice. Int J Med Microbiol. 2021;311:151488. doi: 10.1016/j.ijmm.2021.151488. [DOI] [PubMed] [Google Scholar]
- 171.Kishino S, Takeuchi M, Park S-B, et al. Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc Natl Acad Sci USA. 2013;110:17808–13. doi: 10.1073/pnas.1312937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cani PD, Depommier C, Derrien M, et al. Akkermansia muciniphila: paradigm for next-generation beneficial microorganisms. Nat Rev Gastroenterol Hepatol . 2022;19:625–37. doi: 10.1038/s41575-022-00631-9. [DOI] [PubMed] [Google Scholar]
- 173.Segers A, de Vos WM. Mode of action of Akkermansia muciniphila in the intestinal dialogue: role of extracellular proteins, metabolites and cell envelope components. Microbiome Res Rep . 2023;2:6. doi: 10.20517/mrr.2023.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Facchin S, Bertin L, Bonazzi E, et al. Short-Chain Fatty Acids and Human Health: From Metabolic Pathways to Current Therapeutic Implications. Life (Basel) -> Life (Basel) 2024;14:559. doi: 10.3390/life14050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ryan E, Joyce SA, Clarke DJ. Membrane lipids from gut microbiome-associated bacteria as structural and signalling molecules. Microbiology (Reading) 2023;169:001315. doi: 10.1099/mic.0.001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lamichhane S, Sen P, Alves MA, et al. Linking Gut Microbiome and Lipid Metabolism: Moving beyond Associations. Metabolites. 2021;11:55. doi: 10.3390/metabo11010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cohen LJ, Kang H-S, Chu J, et al. Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist. Proc Natl Acad Sci USA. 2015;112 doi: 10.1073/pnas.1508737112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lynch A, Crowley E, Casey E, et al. The Bacteroidales produce an N-acylated derivative of glycine with both cholesterol-solubilising and hemolytic activity. Sci Rep. 2017;7:13270. doi: 10.1038/s41598-017-13774-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Cohen LJ, Esterhazy D, Kim S-H, et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature New Biol. 2017;549:48–53. doi: 10.1038/nature23874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pérez-Berezo T, Pujo J, Martin P, et al. Identification of an analgesic lipopeptide produced by the probiotic Escherichia coli strain Nissle 1917. Nat Commun. 2017;8:1314. doi: 10.1038/s41467-017-01403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Brown EM, Ke X, Hitchcock D, et al. Bacteroides-Derived Sphingolipids Are Critical for Maintaining Intestinal Homeostasis and Symbiosis. Cell Host Microbe. 2019;25:668–80. doi: 10.1016/j.chom.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wieland Brown LC, Penaranda C, Kashyap PC, et al. Production of α-galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol. 2013;11:e1001610. doi: 10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.An D, Oh SF, Olszak T, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–33. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bae M, Cassilly CD, Liu X, et al. Akkermansia muciniphila phospholipid induces homeostatic immune responses. Nature New Biol. 2022;608:168–73. doi: 10.1038/s41586-022-04985-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Řezanka T, Sigler K. Odd-numbered very-long-chain fatty acids from the microbial, animal and plant kingdoms. Prog Lipid Res. 2009;48:206–38. doi: 10.1016/j.plipres.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 186.An D, Na C, Bielawski J, et al. Membrane sphingolipids as essential molecular signals for Bacteroides survival in the intestine. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4666–71. doi: 10.1073/pnas.1001501107. [DOI] [PMC free article] [PubMed] [Google Scholar]