FIG. 2.
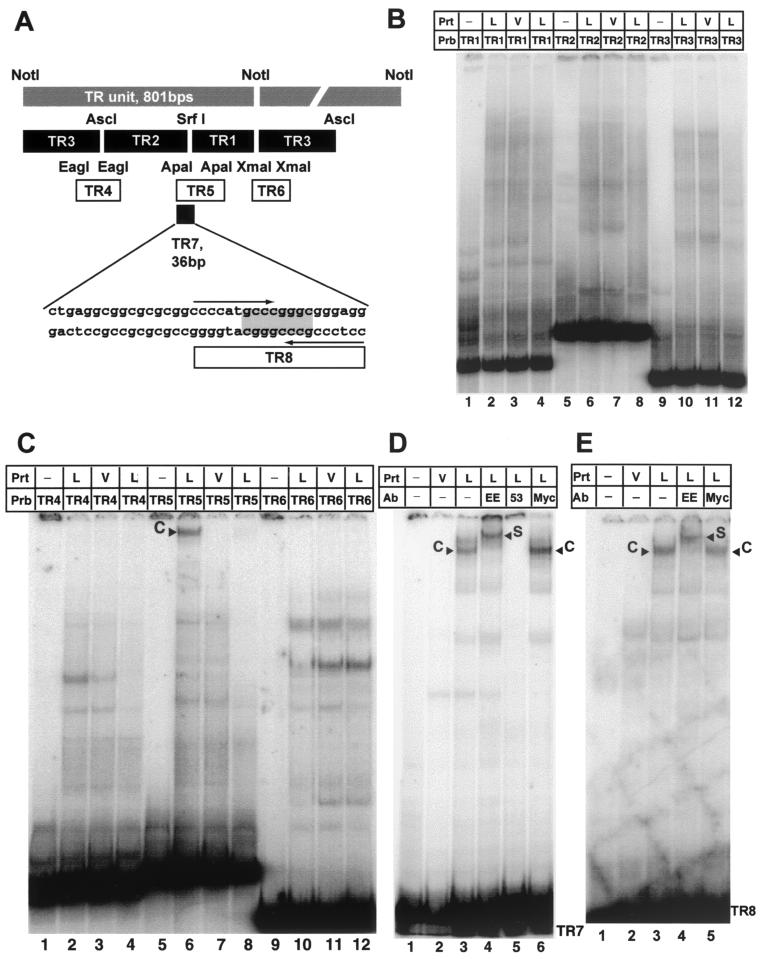
LANA binds specifically to a 20-bp imperfect palindrome within the TR. (A) Diagram of the TR unit as defined by the NotI restriction site (26). Shown are all probes used to identify the LANA binding site within TR7. Plasmids pTR1, pTR2, and pTR3 (kindly provided by M. Lagunoff) were digested with XhoI and NotI to generate the probes TR1, TR2 and TR3, which divide the TR unit into three pieces small enough to be analyzed by EMSA. EagI, ApaI, and XmaI were used to generate fragments TR4, TR5, and TR6, which span the overlapping regions of the first three probes. TR7, 36 bp in length, was created by digestion with ApaI and NarI. The palindromic SrfI site is highlighted; the larger 20-bp imperfect palindrome is indicated by arrows. (B) LANA does not bind to TR1, TR2, and TR3. Probes were radiolabeled with Klenow polymerase and incubated with 4 μg of nuclear extract prepared from MVA/T7-infected CV-1 cells transfected with empty vector (V) or LANA expression vector (L), as indicated above the lanes. Lanes 1, 5, and 9 contain probes (Prb) TR1, TR2, and TR3 in the absence of protein extracts (Prt); lanes 2, 6, and 10 contain protein nuclear extracts of MVA/T7-infected CV-1 cells expressing LANA; lanes 3, 7, and 11 contain extracts from vector-transfected cells. As additional control, lanes 4, 8, and 12 contain LANA and a 50-fold excess of the appropriate unlabeled probe. (C) LANA binds specifically to TR5. Lanes 1, 5, and 9 contain the corresponding probes without protein extracts; lanes 2, 6, and 10 contain protein nuclear extracts CV-1 cells expressing LANA (L); lanes 3, 7, and 11 contain extracts from cells transfected with pEETM-1 vector (V). As a control for specificity, lanes 4, 8, and 12 contain in addition to LANA extract and radiolabeled probe a 50-fold excess of the appropriate unlabeled probe as competitor. Probe TR5 forms a high-molecular-weight complex with LANA extracts (lane 6, labeled C) but not with control extract (lane 7). The observed complex is competed efficiently by the addition of a 50-fold excess of unlabeled probe TR5 as competitor (lane 8). (D) LANA binds specifically to probe TR7 (36 bp in length). Lanes 1 and 2 contain probe TR7 only and control extract from vector-transfected cells (V); lanes 3 to 6 contain protein extract from vaccinia virus-infected CV-1 cells expressing LANA (L). To show that the observed complex is LANA specific, extracts were incubated with three different antibodies (Ab). Lanes 4 and 5 contain EE tag-specific antibody (LANA is tagged at the N terminus [Fig. 1]) or LN53, a LANA-specific monoclonal antibody. Lane 6 contains an unrelated control antibody against a Myc epitope not present in LANA. TR7 forms a complex (labeled C) with LANA-containing extract (lane 3); no complex is seen in lane 2, containing control extract. Lane 4, which contains an antibody to the EE tag, supershifts the complex (labeled S). Lane 5 contains LANA monoclonal antibody LN53, which prevents complex formation. As additional confirmation, unrelated anti-Myc antibody in lane 6 has no effect on complex formation. (E) LANA binds specifically to TR8. Lanes 1 and 2 contain probe TR8 only and control extract from vector-transfected cells (V); lanes 3 to 5 contain protein extract from vaccinia virus-infected CV-1 cells expressing LANA (L); lane 4 contains an antibody specific to the EE tag; lane 5 contains an unrelated control antibody against a Myc epitope not present in LANA. TR8 forms a complex (labeled C) with LANA-containing extract (lane 3); no complex is seen in lane 2, containing control extract. Lane 4, which contains an antibody to the EE tag, supershifts the complex (labeled S).