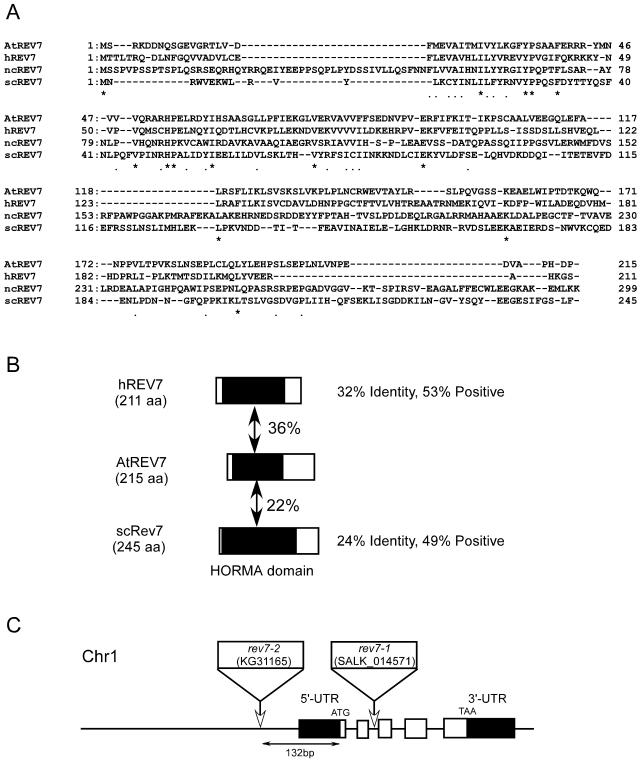
Figure 4.
Alignment of REV7 proteins. A, Alignment of Rev7 proteins from Arabidopsis (AtREV7), human (hREV7), N. crassa (ncREV7), and yeast (scREV7). Amino acid sequences were aligned using Genetyx software (Software Development, Tokyo). Residues that are conserved among all organisms are shown as asterisks (*) and residues that are conserved among more than three kinds of organisms are shown as dots (.). B, Schematic representation of the alignment of hREV7, AtREV7, and yeast REV7 (scREV7). Regions with significant identity are shaded (putative HORMA domain), and percent identity is indicated between AtREV7 and other REV7 proteins. C, Structure of AtREV7 gene and T-DNA insertion site. White boxes represent five exons of the AtREV7 gene and black boxes represent untranslated regions that were supported by the sequence of the expressed sequence tag clone. 5′-UTR and 3′-UTR indicate upstream and downstream, respectively, of the ORF of AtREV7. White rectangles and arrows indicate the T-DNA insertion positions in the upstream region of the ORF (rev7-2) and intron 2 (rev7-1). In rev7-1, left border sequences were found at both sides of the inserted T-DNA. A substitution of three bases was found at the junction of downstream left border and wild-type genome sequence (TGA in Col to AAT in rev7-1). In rev7-2, the T-DNA was accompanied by the insertion of a filler DNA, tcagcgcattttct.