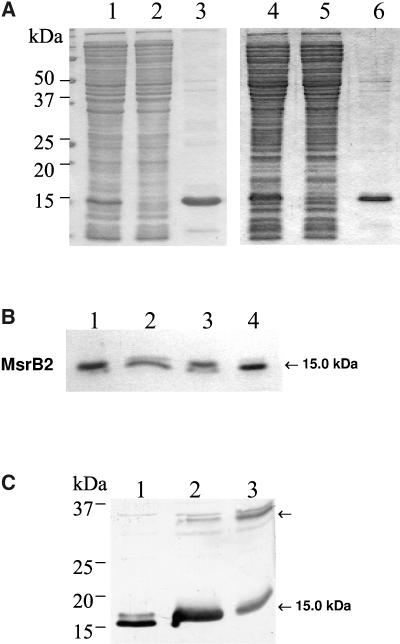
Figure 2.
A, SDS-PAGE analysis of fractions collected during the purification of recombinant MsrB proteins. Lane 1, Crude protein extract of E. coli cells transformed with the pSD80 vector harboring the Arabidopsis MsrB1 gene and induced with 0.1 mm IPTG. Lane 2, Protein extract nonfixated on the Ni affinity-column. Lane 3, MsrB1 protein eluted from the Ni-resin using a buffer containing 500 mm imidazole. Lane 4, Crude protein extract of E. coli cells transformed with the pQE-30 vector harboring the Arabidopsis MsrB2 gene and induced with 0.1 mm IPTG. Lane 5, Similar to lane 2. Lane 6, MsrB2 protein eluted from the Ni-resin using a buffer containing 500 mm imidazole. B, Characterization of the redox state of recombinant Arabidopsis MsrB2. Recombinant Arabidopsis MsrB2 protein was incubated in vitro with reducing reagents (DTE or DTT) or with urea, a denaturing reagent, separated using SDS-PAGE, and revealed by Coomassie Blue. Lane 1, MsrB2; Lane 2, +DTE; Lane 3, +urea 6 m; Lane 4, +DTT. C, SDS-PAGE and western-blotting analysis of recombinant Arabidopsis MsrB2. Crude protein extracts of E. coli cells transformed with the pQE-30 harboring Arabidopsis MsrB2 gene were subjected to SDS-PAGE (13% acrylamide gel), electrotransferred on to a nitrocellulose membrane, and immunoblotted with anti-MsrB polyclonal antibodies. Lane 1, +DTT; Lane 2, −DTT; Lane 3, −DTT, +diamide.