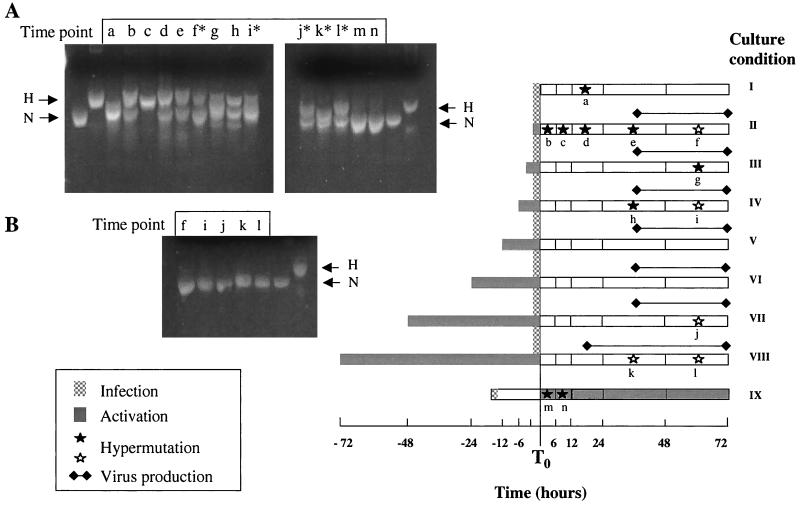
FIG. 6.
Relationship of hypermutation to T-cell activation in virus cultures. CD4+ T cells isolated from the PBMC of normal donors were infected with an NL4-3 virus stock for 2 h at a multiplicity of infection of 0.02 (stippled gray bar) under varying conditions of PHA activation (gray bars). Condition I, infection without addition of PHA; condition II, simultaneous infection and PHA addition; conditions III to VIII, infection with prior PHA activation for 3 to 72 h respectively; condition IX, infection and addition of PHA 24 h later. Cultures were sampled at intervals and assayed for the presence of p24 antigen in the culture supernatant (solid lines) and for the presence of hypermutated HIV-1 DNA sequences. Filled stars, samples where hypermutated sequences were abundant; open stars, samples with rare hypermutated sequences. The appearance of PCR products on HA yellow gels is shown on the right. Time points are labeled “a” through “n” at the tops of the gels and in the diagram. Samples a, b, c, d, e, g, and h are bulk PCR products. Samples from time points at which hypermutation was rare (f, i, j, k, and l) are shown both as bulk PCR products (B) and after pre-enrichment by ScrF1 and AvaII digestion (A). Time points not marked by stars or asterisks yielded only normal sequences both before and after enrichment (not shown).