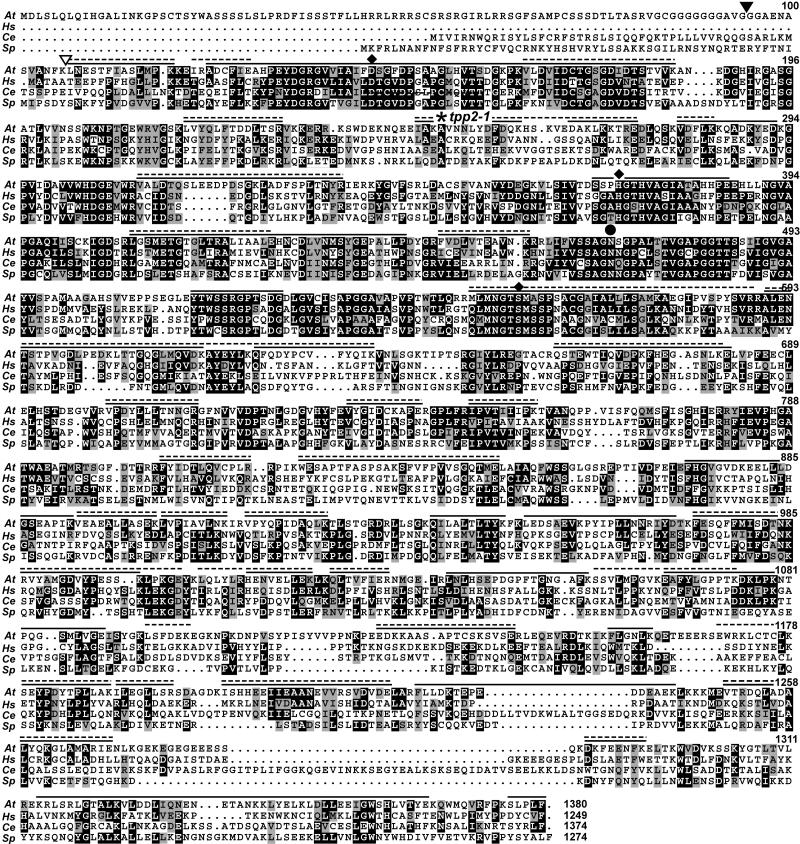
Figure 3.
Amino acid sequence comparison of Arabidopsis TPPII with orthologs from other species. Sequences from Arabidopsis (At), Homo sapiens (Hs), C. elegans (Ce), and S. pombe (Sp) were aligned by ClustalW and viewed with MACBOXSHADE. Identical and similar amino acids are shown in black and gray boxes, respectively. The diamonds identify the conserved Asp, His, and Ser residues that form the catalytic triad. The circle indicates the Asn that stabilizes the tetrahedral intermediate (Renn et al., 1998). The site of the T-DNA insertion for the tpp2-1 mutant is indicated by the asterisk. The solid and dashed lines indicate the composite coverage of the regions from the 153- and 142-kD species, respectively, as identified by MS sequencing of trypsin and Glu-C peptides (see Table II). The black and white triangles locate the N-terminal amino acid of the 153- and 142-kD species, respectively, as determined by Edman degradation of the full-length proteins. GenBank accession numbers for H. sapiens, C. elegans, and S. pombe sequences are NP_003282, NP_495221, and NP_594951, respectively.