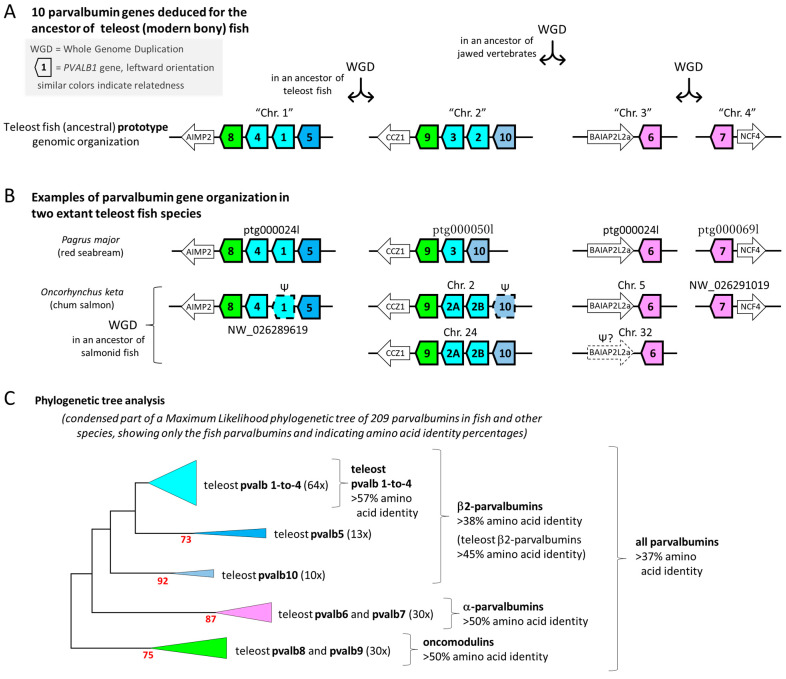
Figure 2.
Parvalbumins in extant teleost fish derive from ten parvalbumin genes in their common ancestor and belong to three different ancient parvalbumin lineages. (A) The immediate ancestor of extant teleost fish possessed at least the genes PVALB1-to-PVALB10, spread over four different chromosomal regions deriving from two whole-genome duplication (WGD) events. The parvalbumin genes are indicated with thick-lined boxes that are pointed in the gene direction and are colored magenta for α-parvalbumins, green for oncomodulins, and different kinds of blue for PVALB1-to-4, ePVALB5, and PVALB10. Neighboring non-parvalbumin genes are indicated by lower boxes with their name abbreviations inside. (B) Parvalbumin gene organization in red seabream and chum salmon, with the direction of the depicted scaffolds adjusted for homogenization. For relevant genomic region information, or Genbank accession numbers providing access to such information, see Supplementary Files S1 and S2. Most symbols are as in (A), and the boxes with dashed lines and Ψ symbols indicate probable pseudogenes. (C) A condensed part of a phylogenetic tree created by the Maximum Likelihood method using 209 parvalbumin amino acid sequences of fishes and other species. Only the teleost fish sequences are indicated here, with between brackets the number of teleost sequences condensed in the respective part of the tree. For the complete tree and sequence information, see [9]. The percentage of trees in which the associated taxa clustered together is shown next to the branches if >50. Percentages of aa identity, calculated with the help of Clustal Omega (https://www.ebi.ac.uk/jdispatcher/msa/clustalo (accessed on 25 March 2024)), are indicated per cluster.