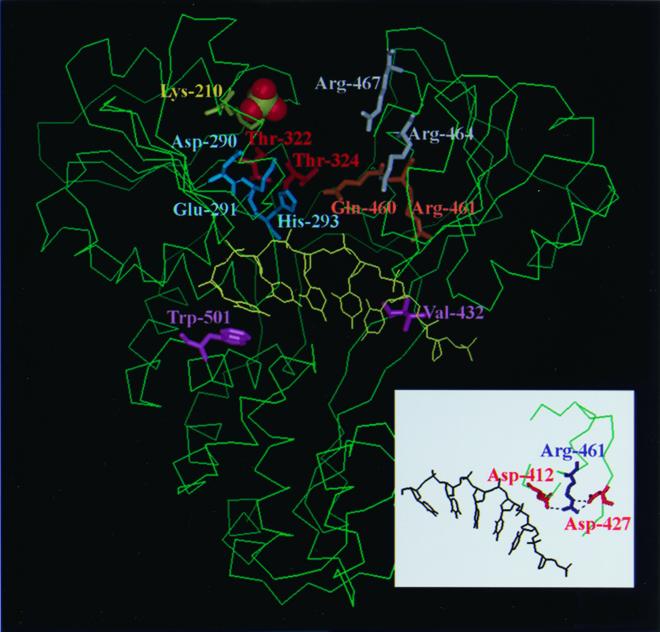
FIG. 5.
Locations of the important amino acid residues in the three-dimensional conformation. The structure of HCV NS3 helicase was based on that determined by Kim et al. (22) using Protein Data Bank (PDB) accession code 1A1V. The important residues identified herein and the Arg-464/Arg-467 previously demonstrated (21, 22, 43) are emphasized by being represented in stick format, while the sulfate ion is represented in Corey-Pauling-Koltun (CPK) format. Residues His-293 and Gln-460 are located in domains I and II, respectively. It is hypothesized that the amine group of the histidine ring of His-293 formed a hydrogen bond with the carbonyl oxygen of the side chain of Gln-460. Residue Arg-461 is located in the interior of domain II. The inset indicates that the Arg-461 can form hydrogen bonds with Asp-412 and Asp-427, which are located at conserved motif V. These figures were drawn using InsightII.