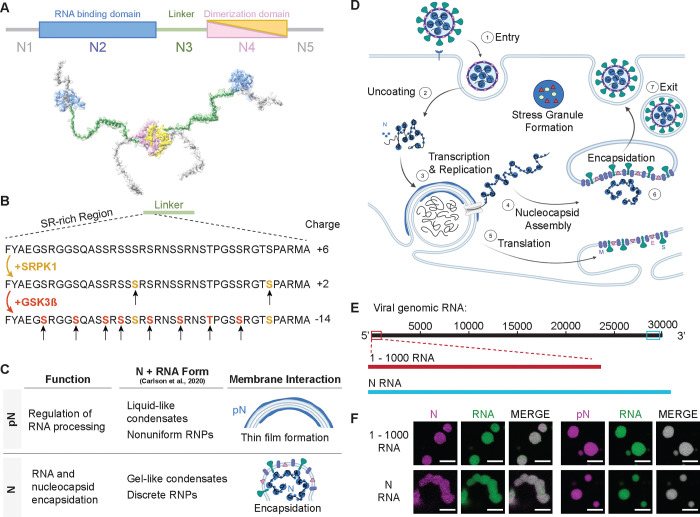
Figure 1: Phosphorylation of the SARS-CoV-2 Nucleocapsid protein (N).
A) Schematic of full-length SARS-CoV-2 Nucleocapsid protein (N) used in experiments, showing N1, a disordered domain; N2, RNA binding domain; N3, disordered linker domain containing the Serine-Arginine-rich (SR) region; N4, dimerization domain; and N5, a disordered domain. B) Sites of phosphorylation in the SR region of the N3 domain. Our experiments used two kinases – SRPK1 and GSK3β – resulting in a maximum of 10 phosphorylation sites. C) Hypothesized form and roles of N and phosphorylated N (pN). pN may have dynamic functions, forms liquid-like droplets with RNA and may localize to the surface of replication organelles. Unmodified N forms solid-like, spherical assemblies with genomic RNA and facilitates new virus encapsulation. N’s form with RNA was characterized by Carlson et al., 2020 using light and electron microscopy. D) A diagram of the SARS-CoV-2 lifecycle. Following viral entry (1), the genome is uncoated (2) such that it can be read. Viral RNA is stored within viral replication organelles (3) wherein RNA transcription and replication may be occurring. N binds to and condenses genomic RNA exiting these organelles (4). In parallel, viral structural proteins are produced (5), in preparation for viral encapsulation (6) and exit (7). E) Main RNA fragments used in experiments are the first 1000 bases from the 5’ end of the viral genome and the 1340 base fragment encoding N. F) Droplets with varied morphologies form upon mixing 40 μM N (5% Alexa Fluor 647 labeled) and 300 nM RNA fragments (5% Cy3 or Cy5 labeled) at 37°C. Scale bar = 5 μm.