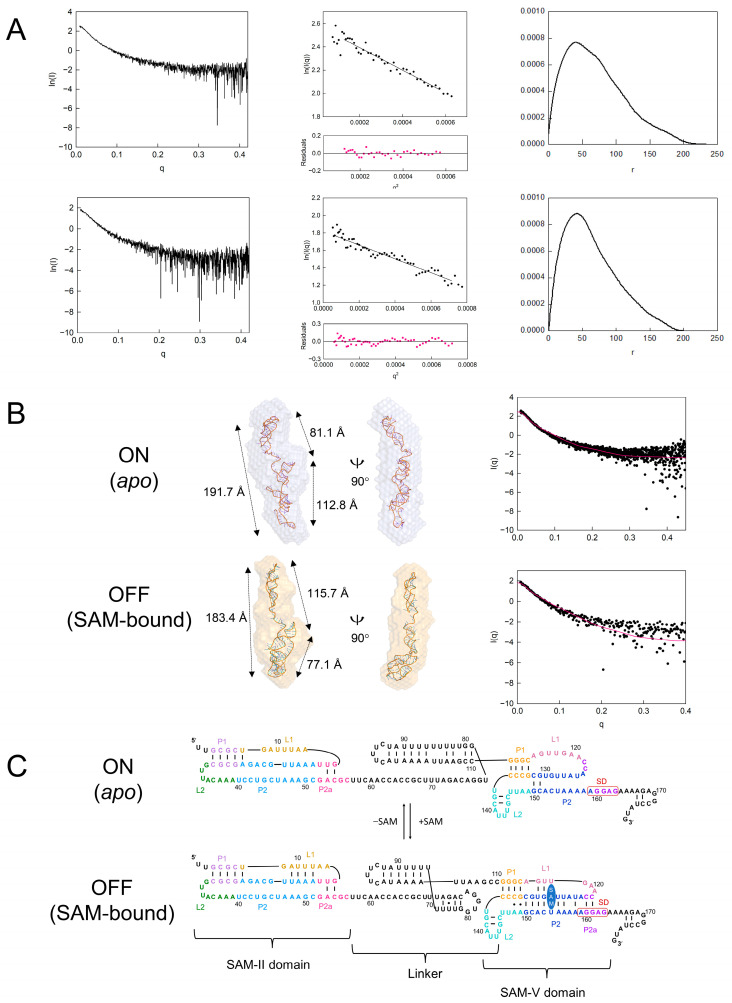
Figure 2.
SAM modulates conformational rearrangements of the full-length tandem SAM-II/SAM-V riboswitch. (A) Scattering profiles (left panel), Guinier plot (middle panel), and pair distribution functions (P (r)) analysis (right panel). The apo (upper row series panels) state measure of the radius of gyration (Rg) is ~59.9 Å and the maximum diameter of the particle (Dmax) is 233. The SAM bound (lower row series panels) measure Rg is ~53.1 Å, and Dmax is 200. (B) Atomic models of the full-length tandem SAM-II/SAM-V riboswitch superimposed with SAXS-derived bead models. The top model for the apo (ON) state is shown in slate; the bottom model for the SAM-bound (OFF) state is shown in bright orange. Atomic model prediction and 3D structure reconstruction were performed using COOT and RNAComposer online servers, which are described in the Material and Methods Section. In the apo (ON) state, the dimensions for the top L2 and bottom tail, top L2 and linker loop, and linker loop and bottom tail are 191.7 Å, 81.1 Å, and 112.8 Å, respectively. In the SAM-bound (OFF) state, the dimensions for the top L2 and bottom tail, top L2 and linker loop, and linker loop and bottom tail are 183.4 Å, 115.7 Å, and 77.1 Å, respectively. The theoretical scattering curve (red) derived from the predicted full-length SAM-II/SAM-V atomic model is mapped with experimental SAXS data plotting by CRYSOL, with apo: χ2 = 2.58 and SAM-bound: χ2 = 2.11. (C) Sequence and secondary structure model of the full-length tandem SAM-II/SAM-V riboswitch studied by SHAPE and small-angle X-ray scattering (SAXS) data. The upper model indicates the apo (ON) state and the lower model indicates the ligand-bound (OFF) state. Coloring in SAM-II and SAM-V domains is consistent with the legend in Figure 1B. The linker is colored black, and the SAM ligand is oval-shaped in blue.