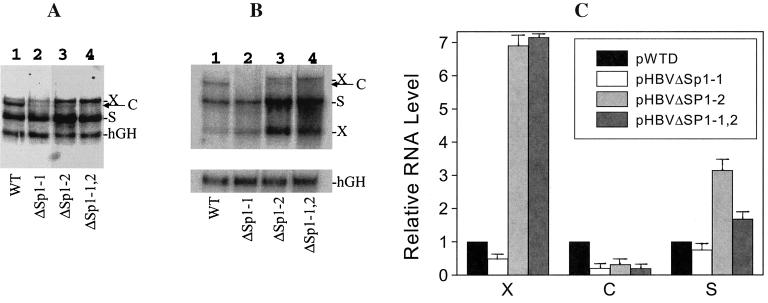
FIG. 3.
(A and B) Northern blot analysis of the HBV RNAs. Huh7 cells transfected with pWTD (lanes 1), pHBVΔSp1-1 (lanes 2), pHBVΔSp1-2 (lanes 3), and pHBVΔSp1-1,2 (lanes 4) genomic DNA constructs were lysed 48 h after transfection for RNA isolation. (A) Northern blot analysis using the 32P-labeled 3.2-kb HBV DNA probe and the hGH cDNA probe together. X, C, and S mark the locations of the 3.9-kb X RNA (13), the C gene transcripts, and the S gene transcripts, respectively. The location of the hGH RNA internal control is also indicated. (B) Northern blot analysis using the 32P-labeled HBV X region-specific DNA probe and the hGH cDNA probe separately. X, C, and S mark the locations of the X, C, and S gene transcripts, respectively. (C) Relative RNA levels expressed by various HBV DNA constructs. X, C, and S indicate the 3.9-kb X gene transcript, the C gene transcripts, and the S gene transcripts, respectively. The RNA bands shown in panel A were measured with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and normalized against the hGH RNA internal control. The X, C, and S RNA levels expressed by the wild-type genome, pWTD, were arbitrarily set as 1. Error bars indicate standard deviations.