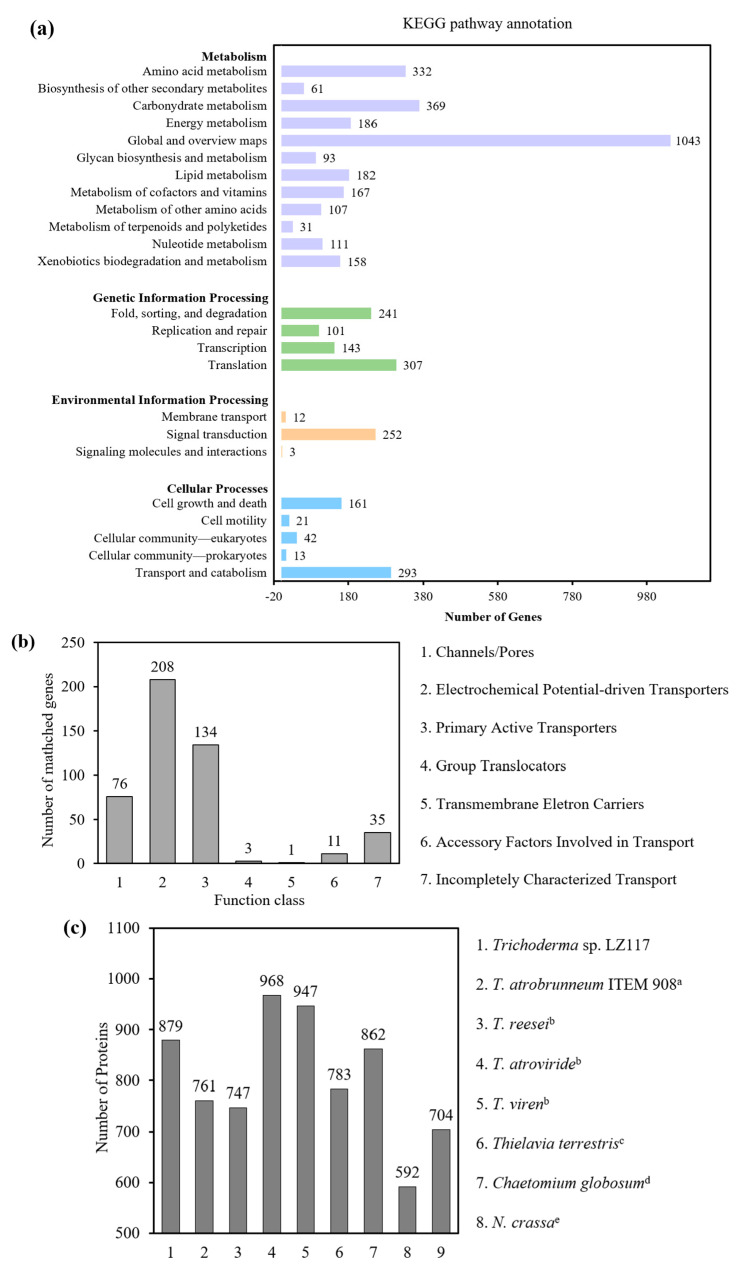
Figure 4.
Functional annotation of Trichoderma sp. LZ117. (a) KEGG functional annotation of Trichoderma sp. LZ117. The y-axis indicates the enriched pathway names, and the x-axis denotes the number of coding genes in the enriched pathways. (b) TCDB functional annotation of Trichoderma sp. LZ117. The y-axis shows the number of matched coding genes, and the x-axis denotes the name of the function class. (c) Prediction of secreted proteins in Trichoderma sp. LZ117 and other Trichoderma spp. The y-axis indicates the number of the predicted proteins, and the x-axis denotes the name of the selected Trichoderma spp. a Data from Fanelli F, Liuzzi VC, et al., 2018 [10]. b Data from Druzhinina IS, Shelest E, et al., 2012 [48]. c Data from Berka RM, Grigoriev IV, et al., 2011 [49]. d Data from Cuomo CA, Untereiner WA, et al., 2015 [50]. e Data from Galagan JE, Calvo SE, et al., 2003 [51]. f Data from Galagan JE, Calvo SE, et al., 2005 [52].