Figure 2.
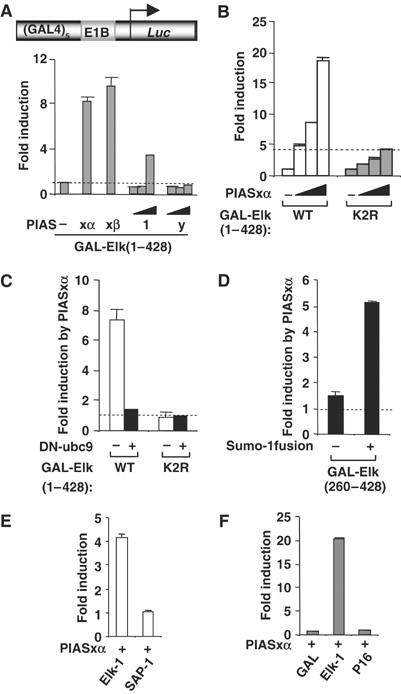
Activation of sumoylated Elk-1 by PIASxα. Reporter gene analysis of the activities of GAL-Elk fusion proteins on the GAL-driven E1b promoter-reporter construct (schematic shown in [A]) in 293 cells is shown. (A) The activity of GAL-Elk(1–428) was tested in the presence and absence of cotransfected PIASxα (0.1 μg), PIASxβ (0.1 μg), PIAS1 (0, 0.1, 0.5 and 1 μg) and PIASy (0, 0.1, 0.5 and 1 μg) as indicated and presented as fold induction of reporter activity relative to the absence of PIAS proteins. (B) The activities of the wild-type (WT) and mutant (K2R) GAL-Elk(1–428) fusion proteins were tested in the presence or absence of increasing amounts of PIASxα (0, 10, 25 and 100 ng) as indicated and presented as fold induction of reporter activity relative to the absence of PIASxα (taken as 1). For alternative depiction of data, see Supplementary Figure S1. (C) The activities of GAL-Elk(1–428) WT and K2R were tested in the presence or absence of PIASxα (0.25 μg) and/or DN-ubc9 (1 μg) as indicated. The data are presented as fold induction by PIASxα of reporter activity and are calculated from the activity of the reporter under each condition relative to the activity in the absence of PIASxα. (D) The activities of GAL-Elk(260–428) and GAL-Sumo-Elk(260–428) were tested in the presence or absence of PIASxα (0.25 μg). The data are presented as fold induction by PIASxα of reporter activity and are relative to the activity of each construct in the absence of PIASxα. (E, F) Activities of GAL-Elk(223–428), GAL-SAP-1(234–431) (E), GAL-DBD, GAL-Elk(1–428) and GAL-VP16 (F) in 293 cells in the presence or absence of transfected PIASxα. Data are shown as fold induction relative to the activity of each GAL fusion in the absence of PIASxα. Luciferase assays are representative of at least two independent experiments (standard errors are shown; n=2).
