Figure 6.
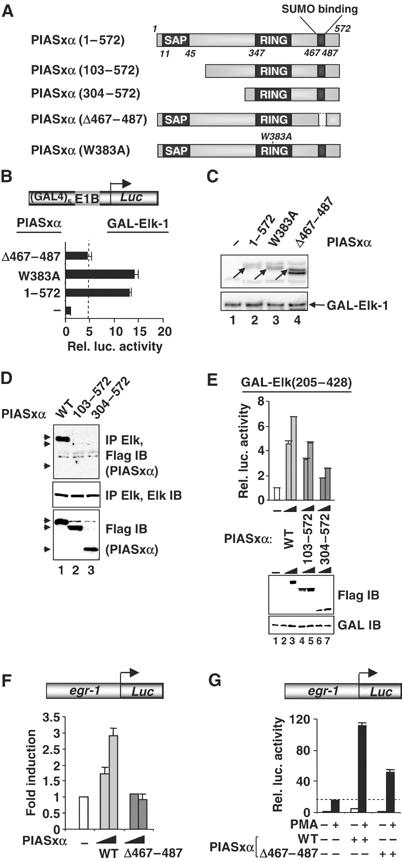
Mapping the determinants in PIASxα for coactivation of Elk-1. (A) Schematic of PIASxα proteins used in this figure. The known PIAS domains (SAP, RING and SUMO binding) are indicated in dark grey boxes. (B) Reporter gene analysis of activities of GAL-Elk(1–428) in the presence or absence of the indicated PIASxα proteins on the GAL-driven E1b promoter-reporter construct in 293 cells. The data are shown relative to GAL-Elk(1–428) in the absence of PIASxα. (C) Western blots probed with Flag (PIASxα) and Gal4 (GAL-Elk) antibodies. Arrows indicate the band corresponding to the protein with the correct size. (D) Co-immunoprecipitation analysis of the indicated PIASxα deletion mutants and cotransfected Elk-1 using an anti-Elk-1 antibody. Arrows represent the locations of the truncated PIASxα proteins. (E) Reporter gene analysis of increasing amounts of the indicated PIASxα proteins (wild type (WT), 125 and 250 ng; 103–572, 500 and 1000 ng; 304–572, 750 and 1500 ng) and the Elk-1 derivative GAL-Elk(205–428) on the GAL-driven E1b promoter-reporter construct in 293 cells. Expression levels of the mutants are shown in the accompanying Western blot. (F, G) Reporter gene analysis of activities of egr-1 promoter-driven luciferase reporter construct in 293 cells. (F) Reporter activity in the absence (white bar) or presence of increasing amounts of WT (light grey bars) or mutant (Δ467–487) (0.1 and 1 μg) (dark grey bars) PIASxα proteins (relative to the basal level of the reporter construct). (G) Reporter activities in the absence (white bar) or presence of WT or mutant (Δ467–487) (dark grey bars) PIASxα proteins in the presence or absence of PMA. The data are shown relative to the basal level of the reporter construct (taken as 1). Luciferase assays are representative of at least two independent experiments (standard errors are shown; n=2).
