Figure 2.
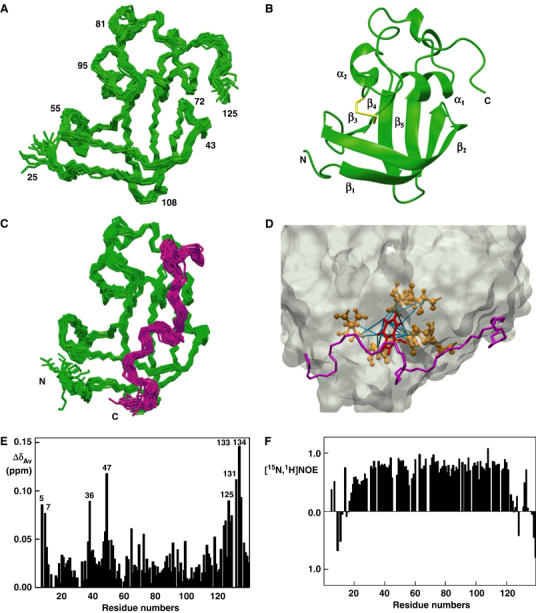
NMR studies on FimDN. (A) Polypeptide backbone of FimDN(25–125) represented by a bundle of 20 energy-minimized DYANA conformers. Selected positions along the polypeptide chain are identified with sequence positions. (B) Ribbon drawing of one of the 20 energy-minimized conformers. β1–β5 and α1–α2 indicate five β-strands and two α-helices, respectively. The disulfide bridge Cys63–Cys90 is drawn in yellow. The chain ends are identified by the letters N and C. (C) NMR structure of FimDN(25–139) represented by a bundle of 20 energy-minimized DYANA conformers showing only the polypeptide backbone. The chain ends are identified by the letters N and C. The C-terminal residues 125–139 are shown in magenta. (D) Close-up view of the surface of one of the 20 energy-minimized conformers of FimDN(25–139). Relative to (C), the structure has been rotated by approximately 90° about a vertical axis. The backbone of the C-terminal stretch 125–139 is drawn in magenta, and the side chain of Trp133 is indicated in red. Those side chains which show long-range NOE connectivities with Trp133 are drawn in bronze. In total, 14 long-range upper-distance limits between Trp133 and the rest of the protein (shown in cyan) define the position of the aromatic ring of Trp133. (E) Chemical shift variations of FimDN upon binding to FimC–FimHP. ΔδAv is the weighted average of the 15N and 1H chemical shifts, 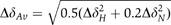 (Pellecchia et al, 1999). (F) Heteronuclear [15N,1H]NOE measurements of FimDN(1–139) in the FimDN–FimC–FimHP ternary complex. Values between 0.5 and 1 indicate well-structured parts of the protein; values<0.5 manifest increased flexibility.
(Pellecchia et al, 1999). (F) Heteronuclear [15N,1H]NOE measurements of FimDN(1–139) in the FimDN–FimC–FimHP ternary complex. Values between 0.5 and 1 indicate well-structured parts of the protein; values<0.5 manifest increased flexibility.
