Abstract
Haemaphysalis longicornis (Neumann), the Asian longhorned tick, is a species native to East Asia, but invasive to Australia, New Zealand, and most recently, the United States. It has spread rapidly across the eastern United States after being established in New Jersey in 2017. Aiding this rapid expansion is the ability of this tick to reproduce parthenogenically and feed on diverse host species. In cattle, this tick can cause heavy burdens and act as a vector for the pathogenic hemoprotozoan parasite Theileria orientalis, genotype Ikeda, creating economic losses that impact the cattle industry. Here, we report Asian longhorned ticks, collected from cattle, a dog, and pastures and morphologically identified at the Oklahoma Animal Disease Diagnostic Laboratory as H. longicornis before molecular confirmation through PCR amplification of the cox1 gene. Blood samples from infested cattle were collected and assessed molecularly for the presence of T. orientalis, with no pathogenic DNA detected. This report describes the first record of H. longicornis in Oklahoma and the farthest westward detection of this tick in the United States to date.
Keywords: Haemaphysalis longicornis, cattle, Theileria orientalis, invasive species, range expansion, Oklahoma
1. Introduction
Detection of Haemaphysalis longicornis (Neumann, 1901) in 2017 on an Icelandic sheep in New Jersey prompted widespread investigation of this invasive species, where a subsequent review of research collections revealed the earliest known non-quarantined introduction of this species to the United States occurred in 2010 [1,2]. Since 2017, surveillance predominantly in the eastern United States has detected H. longicornis in 20 states, including Washington D.C. [3]. Prior to the present report, this tick has been detected as far west as Arkansas and as far south as Georgia; as such, it is primarily distributed within the east and northeastern regions of the United States [3]. Asian longhorned ticks follow a three-host life cycle and have been collected from a wide variety of mammal and avian hosts in the United States, including companion animals, livestock, wildlife, and humans. Heavy infestation burdens have been documented on multiple species, notably cattle and white-tailed deer (Odocoileus virginianus) [4]. Host-seeking ticks have been frequently collected from the environment within the United States, including in both grassy pastures and wooded areas [4].
The rapid range expansion of this tick following its reported introduction is accelerated by multiple factors, including the species’ unique parthenogenic characteristic, ability to feed on a variety of host species, and survivability in a range of environmental conditions [5,6,7]. Some populations of H. longicornis have been found to tolerate temperatures between −2 °C and 40 °C and can be found in multiple habitats including pastures and mixed hardwood forests [8,9].
Molecular investigation of specimens collected in the United States revealed a closer genetic consistency with parthenogenic populations rather than bisexual populations of H. longicornis in other regions of the world, which is supported by the general lack of male specimens found in the US [5]. In places where parthenogenic ticks predominate, triploid females can reproduce without the presence of males by producing genetic clones, quickly building dense focal populations [10]. Haplotyping based on sequences of the mitochondrial cox1 gene reveals that the Asian longhorned tick invasion of the United States originated from East Asian populations rather than Australian populations [5,11]. Ticks found within the US since 2017 are consistent with haplotypes H1, H2, and H3, which are commonly found in China and indicate the potential for three separate introductions into the country [5].
Worldwide, Haemaphysalis longicornis is traditionally considered a nuisance to livestock, and the major vector for the hemoparasite Theileria orientalis. Currently, eleven genotypes of T. orientalis have been identified according to variability in the major piroplasm surface protein (MPSP) [12,13]. Before the introduction of H. longicornis to the US, several T. orientalis genotypes had been identified, including Chitose and Buffeli, both usually considered non-pathogenic [14,15]. However, after detecting H. longicornis in the US, an increase in clinical bovine theileriosis cases has been reported [15,16,17]. This bloodborne parasite is transmitted to cattle mainly during tick feeding, where the organism infects erythrocytes and leukocytes, resulting in anemia, pyrexia, jaundice, weakness, and potentially death [18]. Infected cattle have been reported primarily in Virginia and West Virginia, where cattle populations are less dense than in South Central states, such as Oklahoma [16,19]. As this tick expands into areas with higher livestock densities, the impact on the health of the agricultural industry due to the presence of this tick and pathogen may become more prominent.
In other countries, Asian longhorned ticks can transmit numerous bacterial, protozoal, and viral pathogens of veterinary and medical importance [20]. Concern within the United States regarding H. longicornis as a potential vector for numerous endemic pathogens of public health and veterinary importance has arisen as their range continues to expand into regions where these pathogens are vectored by other tick species. Multiple pathogens endemic to the United States have been detected in H. longicornis, including Anaplasma phagocytophilum, Ehrlichia spp., and Rickettsia spp., although vector competency has been largely undetermined for many of these pathogens [21,22,23]. Acquisition of Rickettsia rickettsii by co-fed H. longicornis has been observed experimentally, but evidence of natural acquisition or transmission has not been shown [24].
The present report documents the first collection of H. longicornis within the state of Oklahoma and identifies the first counties with established populations of this species in the state. This report will provide further information regarding the source of collection and the potential impact of this species as its range expands into the South Central United States.
2. Materials and Methods
2.1. Ticks
2.1.1. Tick Submission and Morphologic Identification
On 31 July 2024, a local veterinarian reported several heifers from Craig County, Oklahoma, with a heavy tick burden. The ticks appeared different from the species typically seen in the area before. A total of 13 engorged ticks removed from cattle were submitted to the Oklahoma Animal Disease Diagnostic lab for identification. A second tick submission on 9 August 2024, from the same ranch but in Mayes County, located adjacent to Craig County, OK, included ticks removed from cattle and a dog and passively collected from pasture (N = 20). Ticks were submitted in 2 individual tubes with 70% ethanol, but not individually identified by source. Morphologic identification was performed at the Oklahoma Animal Disease Diagnostic Laboratory through parasitology, using stereomicroscopy and dichotomous keys [25,26].
2.1.2. DNA Extraction
A subset of identified ticks, consisting of 2 engorged specimens collected from cattle (labeled OK3, OK4) and 2 unfed environmentally collected specimens (labeled OK5, OK6) were individually washed in 3% hydrogen peroxide, 70% alcohol, and a phosphate-buffered solution (PBS) sequentially. Washed ticks were manually dissected by circumferentially incising the idiosoma and removing all internal contents using sterile scalpel blades before the extraction of total genomic material using the Quick-DNA Miniprep Plus Kit (Zymo Research, Irvine, CA, USA), following manufacturer instructions. An extraction control utilizing water was made to ensure no contamination occurred during DNA isolation. The remaining exoskeletons were stored in ethanol at 20 °C.
2.1.3. Amplification of cox1 Gene
Extracted genomic material was used to amplify a 710 bp fragment of the mitochondrial cytochrome c oxygenase gene (cox1) by PCR using primers LCO1490-5′-GGTCAACAAATCATAAAGATATTGG-3’ and HCO2198-5′-TAAACTTCAGGGTGACCAAAAAATCA-3′ which have been previously used for cox1 haplotyping of Haemaphysalis longicornis [5,27]. Individual 50 µL reactions included 25 µL of GoTaq DNA polymerase (Promega, Madison, WI, USA), 1 µL of each primer, 1 µL of bovine serum albumin (BSA), 5 µL of sample DNA, and 17 µL of nuclease-free water. Thermocycler conditions were as follows: initial denaturation at 95 °C for 3 min, and then 40 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 30 s, and extension at 72 °C for 1 min before a final extension at 72 °C for 7 min. Appropriate negative controls were used. All PCR products were resolved by gel electrophoresis in 1.5% agarose gels and observed using GelRed Nucleic Acid Stain (Biotum, Fremont, CA, USA). Positive amplicons were purified using the GeneJET genomic DNA purification kit (Thermo Fischer, Waltham, MA, USA) according to the manufacturer’s instructions before bidirectional Sanger sequencing at Oklahoma State University (Stillwater, OK, USA).
2.1.4. Phylogenetic Analysis
The resulting sequence data were evaluated and aligned using Geneious Prime Software V.2024.05 (Dotmatics, Boston, MA, USA) to create 612 bp consensus sequences of each tick for species confirmation through comparison with previously published reference sequences of H. longicornis, accessible in the Nucleotide BLAST database (National Library of Medicine). Sequence data from the 4 extracted Oklahoma ticks, as well as reference sequences in GenBank representing the 3 known haplotypes of H. longicornis (accession numbers MT034323, MT034173, and MT034418) currently documented within the United States, were used to generate a Neighbor-Joining phylogenetic tree to visualize the haplotype grouping of the presently collected ticks. Sequences derived from ticks in the present study are available in the GenBank database under accession numbers PQ231312–PQ231315.
2.2. Theileria orientalis
Due to concerns for the presence of T. orientalis, genotype Ikeda, in Oklahoma cattle, screening for this protozoan parasite started in November 2022, when a cattle herd from Lincoln County, OK, reported animals having clinical signs similar to Anaplasma marginale, despite testing negative for this pathogen. Even though H. longicornis was not reported in 2022, the movement of animals and the lack of detection of ticks were initially a concern.
2.2.1. Cattle Blood Samples
On 9 August 2024, a total of 10 blood samples from cattle originally identified as infested with H. longicornis were collected in EDTA tubes to be tested for T. orientalis.
Additionally, before the detection of H. longicornis in Oklahoma from November 2022 to June 2024, 179 blood samples from cattle in Oklahoma were tested for T. orientalis due to concerns about the presence of the Ikeda genotype in the area.
2.2.2. DNA Extraction
DNA was isolated from whole blood samples using the ReliaPrep Blood gDNA Miniprep System (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
2.2.3. Amplification of MPSP and SSU rRNA Genes
We amplified the MPSP by utilizing the primers MPSP-F (5′-CTTTGCCTAGGATACTTCCT-3′) and MPSP-R (5′-ACGGCAAGTGGTGAGAACT-3ing′) and utilizing the same conditions for the H. longicornis cox1 PCR as described above but with a melting temperature of 57 °C for 1 min. Additionally, we amplified the small subunit (SSU) rRNA gene using primers SSU-F (5′-ATTGGAGGGCAAGTCTGGTG-3′) and SSU-R (5′-CTCTCGGCCAAGGATAAACTCG-3′) with the same conditions as for the MPSP gene [28,29]. Reactions were conducted on all 10 blood samples from Oklahoma cattle infested with H. longicornis and all 4 tick specimens extracted for the present report. Additionally, the same testing was performed on the 170 blood samples from Oklahoma cattle that were collected prior to detecting H. longicornis in the state. All positive amplicons were purified as described above and submitted for Sanger sequencing to Eurofins Genomics (Louisville, KY, USA). Due to the similarity of clinical signs observed in animals infected with Anaplasma marginale to those infected with T. orientalis, a PCR that amplifies a fragment of the MSP4 gene was performed, utilizing the primers MSP45 (5′-GGGAGCTCCTATGAATTACAGAGAATTGTTTAC-3′) and MSP43 (5′-CCGGATCCTTAGCTGAACAGGAATCTTGC-3′) on all bovine blood samples as described by de la Fuente et al., 2003 [30].
3. Results
3.1. Tick Morphologic Identification
All ticks submitted were morphologically identified as H. longicornis adult females [25,26] (Figure 1 and Figure 2: images captured using the Olympus cellSens Entry imaging software version 3.2 and an Olympus model SZX2-ILLTQ microscope, Olympus Life Science, Waltham, MA, USA).
Figure 1.
Dorsal (A) view and ventral (B) view of an engorged female H. longicornis identified after collection from Oklahoma cattle. The ticks pictured measure approximately 5 mm in width by 8 mm in length. The characteristic lateral projection on palpal segment 2 (black arrowheads) and a dorsal spur on palpal segment 3 can be seen.
Figure 2.
Multiple engorged female H. longicornis identified after collection from cattle residing in Oklahoma.
3.2. Tick Molecular Identification
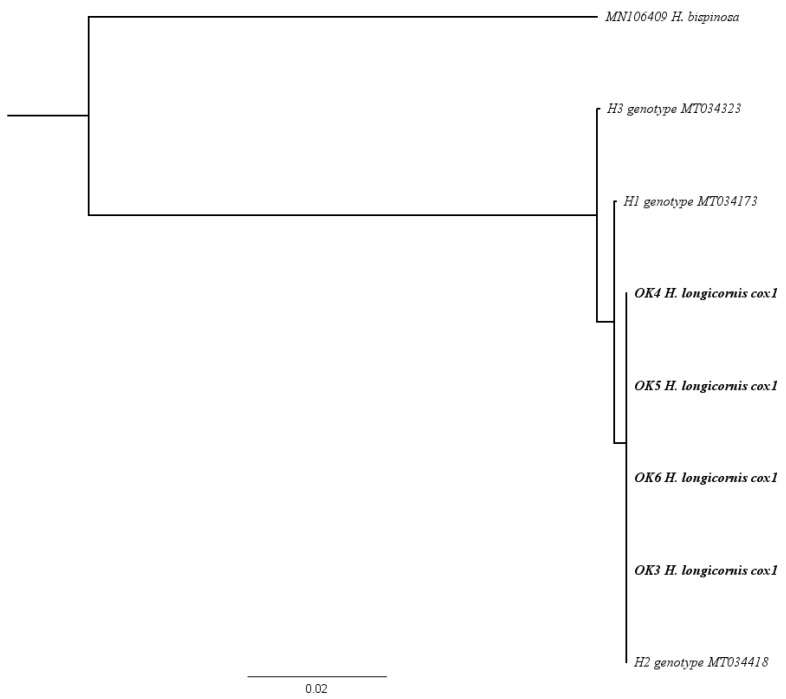
Genomic sequences derived from the four extracted ticks were 100% consistent with previously published cox1 sequences of H. longicornis within the United States. All four extracted ticks from the present report were 100% identical to each other. Phylogenetic analysis based on a Neighbor-Joining phylogenetic tree and gene comparison using Nucleotide BLAST from the National Center for Biotechnology Information database (GenBank) determined all cox1 sequences from the present report to be 100% identical to a previously submitted cox1 sequence identified as haplotype 2 (MT034418), the predominant haplotype present in the United States (Figure 3). Only one or two base pairs differed between the Oklahoma ticks and genotypes H1 (MT034173) and H3 (MT03423).
Figure 3.
Neighbor-Joining phylogenetic tree depicting the relationship between cox1 sequences of haplotypes previously documented in the United States (H1, H2, H3) and ticks collected for the present report (shown in bold), out-grouped to Haemaphysalis bispinosa. Ticks from this study are designated with OK3–OK6.
3.3. Theileria orientalis
None of the blood samples collected from cattle (N = 10) infested with H. longicornis or individual H. longicornis ticks (N = 4) in Mayes and Craig counties, OK, were detected as infected with T. orientalis.
From the surveillance samples prior to the H. longicornis finding in Oklahoma, a total of 11/170 (6.1%) were positive for T. orientalis, genotype Chitose, and 6 were positive for Anaplasma marginale 6/170 (3.3%); GenBank accession numbers are PQ300672 and PQ301201 for the MPSP and SSU rRNA genes, respectively (Table 1).
Table 1.
Number of bovine blood samples tested positive for Anaplasma marginale or Theileria orientalis by PCR according to Oklahoma county and year of collection.
| Year Collected | County (OK) |
A.marginale PCR Positive |
T. orientalis PCR Positive * |
Total Tested |
|---|---|---|---|---|
| 2022 | Atoka | 0 | 3 | 61 |
| 2023 | Creek | 0 | 0 | 4 |
| 2022–2023 | Lincoln | 0 | 4 | 49 |
| 2024 | Mayes | 0 | 0 | 10 |
| 2022–2023 | Okfuskee | 0 | 4 | 43 |
| 2022 | Pawnee | 1 | 0 | 1 |
| 2022 | Payne | 4 | 0 | 10 |
| 2024 | Pushmataha | 0 | 0 | 1 |
| 2022 | Washington | 1 | 0 | 1 |
* Theileria orientalis genotype Chitose detected.
4. Discussion
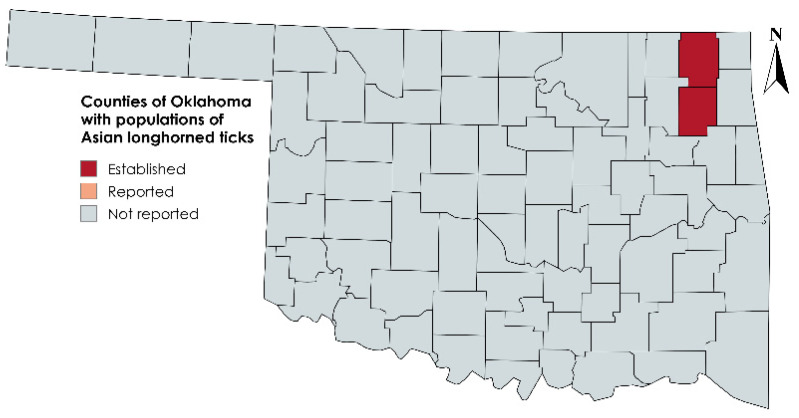
In the present report, we describe the first discovery of H. longicornis parasitizing cattle in Oklahoma, collected in pastures where cattle reside. Despite active and ongoing surveillance efforts, the presence of H. longicornis in Oklahoma has not been previously described to the authors’ knowledge; the identification of these ticks confirms their presence in the state and extends the westward border of their known range within the United States. Based on the standards for establishing tick distributions and abundance set by the CDC, the ticks collected for the present report indicate that Craig and Mayes counties in northeast Oklahoma have an established population of H. longicornis, the first documented within the state (Figure 4) [31]. The presence of this tick in Oklahoma can be expected to have an impact on the cattle industry as well as on public health if it continues to expand within the state.
Figure 4.
Map of Oklahoma depicting counties with recently established populations of Asian longhorned ticks. Established is defined as the identification of at least six individual ticks, or at least two of three host-seeking life stages within one calendar year. Reported is defined as the identification of less than six individual ticks of a single life stage within one calendar year [31].
Oklahoma continually ranks within the top five states for inventory of all cattle and calves within the United States, with a reported 4.60 million heads, representing 5.1% of all cattle in the country [32]. In numbers of beef cattle alone, Oklahoma ranks second in the country behind neighboring Texas. In addition to Oklahoma, the top four overall cattle-producing states include primarily those along historic cattle drives (i.e., Texas, Nebraska, and Kansas), many of which are threatened by the range expansion of this tick based on multiple predictive models [6,33]. The expansion of H. longicornis and T. orientalis Ikeda into the region threatens to burden the cattle industry by increasing animal mortality and decreasing overall production in areas with a density of animals not yet encountered within the current range of H. longicornis in the United States. States such as Virginia, which reports the highest number of counties with established H. longicornis populations at 39 counties and has documented cases of T. orientalis Ikeda, reports a 1.3 million head total of cattle and calves, less than half of that of Oklahoma [16,34]. Within the United States, limited data are available regarding the economic impact of H. longicornis and T. orientalis; however, marked economic loss has been reported in Asia, New Zealand, and Australia from clinical bovine theileriosis, with an estimated AUD 20 million annual loss reported in Australia alone [18,35].
Parasitism of cattle by ticks may cause physical damage, anemia, and “tick worrying”, resulting in detrimental irritation and unrest, and the transmission of pathogens [36]. These can result in the decreased production and well-being of animals and, in some cases, may result in death. Outbreaks of clinical theileriosis in beef and dairy cattle may result in significant losses to production and increased mortality [37,38]. Fatalities in US cattle that were heavily parasitized by H. longicornis have been described [1]. While H. longicornis has been determined to be an incompetent vector for Babesia bovis, a causative agent of Texas Cattle Fever, other pathogens including Heartland virus and Bourbon virus may be vectored by this tick, indicating the potential for an important One Health implication beyond the cattle industry [39,40,41]. Despite its association as a parasite of cattle, H. longicornis will readily feed on other domestic species and humans, so attention to potential human pathogens vectored by this tick is crucial. Outside of the United States, pathogens infecting other host species including Babesia gibsoni in dogs can be vectored by H. longicornis, indicating the potential for spreading within the United States with a competent vector present. However, experimental evidence has yet to be seen in US populations [42,43].
Management strategies on infested livestock operations in Tennessee that showed promise in reducing H. longicornis populations included implementing active surveillance methods, appropriate use of on-animal acaricides, pasture management, and maintenance of closed herds. Implementation of multiple of these strategies was found to have significantly reduced the questing of H. longicornis collected over 2 years [44]. Adoption of these strategies may be useful for Oklahoma livestock producers with the presence of H. longicornis populations. Animals with clinical signs consistent with anaplasmosis or theileriosis should be tested for both A. marginale and T. orientalis as the prevalence of this tick expands.
5. Conclusions
The present report describes the first case of H. longicornis in Oklahoma and identifies Craig County as the first county with an established population of these ticks within the state, followed by Mayes County. With the anticipated spreading of this tick in the state, continued active tick surveillance and testing of identified ticks and cattle for T. orientalis remains a priority. Increasing public awareness about H. longicornis and the potential pathogens it may transmit to people, pets, and livestock is important to reduce the threat to public health and animal husbandry in Oklahoma.
Acknowledgments
We would like to thank Rod Hall (DVM, Oklahoma Department of Agriculture and Forestry) and Zeke Proctor (DVM, USDA-APHIS) for submitting and collecting the ticks identified in this report and Tyler Thomas (DVM) for collecting the blood samples from cattle before the identification of H. longicornis in the state of Oklahoma.
Institutional Review Board Statement
Due to samples being collected and submitted by veterinarians for diagnosis and identification, ethical review and approval were waived for this study.
Data Availability Statement
Data from this study are freely available within the GenBank database under accession numbers PQ231312–PQ231315; PQ300672; and PQ301201.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Rainey T., Occi J.L., Robbins R.G., Egizi A. Discovery of Haemaphysalis longicornis (Ixodida: Ixodidae) Parasitizing a Sheep in New Jersey, United States. J. Med. Entomol. 2018;55:757–759. doi: 10.1093/jme/tjy006. [DOI] [PubMed] [Google Scholar]
- 2.Beard C.B. Multistate Infestation with the Exotic Disease–Vector Tick Haemaphysalis longicornis—United States, August 2017–September 2018. Morb. Mortal. Wkly. 2018;67:1310–1313. doi: 10.15585/mmwr.mm6747a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.USDA, APHIS National Haemaphysalis longicornis (Asian Longhorned Tick) Situation Report. [(accessed on 19 August 2024)];2024 Available online: https://www.aphis.usda.gov/sites/default/files/monitoring-h-longicornis-plan-final-january-2024.pdf.
- 4.Thompson A.T., White S.A., Doub E.E., Sharma P., Frierson K., Dominguez K., Shaw D., Weaver D., Vigil S.L., Bonilla D.L., et al. The wild life of ticks: Using passive surveillance to determine the distribution and wildlife host range of ticks and the exotic Haemaphysalis longicornis, 2010–2021. Parasit. Vectors. 2022;15:331. doi: 10.1186/s13071-022-05425-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egizi A., Bulaga-Seraphin L., Alt E., Bajwa W.I., Bernick J., Bickerton M., Campbell S.R., Connally N., Doi K., Falco R.C., et al. First glimpse into the origin and spread of the Asian longhorned tick, Haemaphysalis longicornis, in the United States. Zoonoses Public Heallth. 2020;67:637–650. doi: 10.1111/zph.12743. [DOI] [PubMed] [Google Scholar]
- 6.Rochlin I. Modeling the Asian Longhorned Tick (Acari: Ixodidae) Suitable Habitat in North America. J. Med. Entomol. 2019;56:384–391. doi: 10.1093/jme/tjy210. [DOI] [PubMed] [Google Scholar]
- 7.Cumbie A.N., Whitlow A.M., Arneson A., Du Z., Eastwood G. The Distribution, Seasonal Abundance, and Environmental Factors Contributing to the Presence of the Asian Longhorned Tick (Haemaphysalis longicornis, Acari: Ixodidae) in Central Appalachian Virginia. J. Med. Entomol. 2022;59:1443–1450. doi: 10.1093/jme/tjac067. [DOI] [PubMed] [Google Scholar]
- 8.Tufts D.M., VanAcker M.C., Fernandez M.P., DeNicola A., Egizi A., Diuk-Wasser M.A. Distribution, Host-Seeking Phenology, and Host and Habitat Associations of Haemaphysalis longicornis Ticks, Staten Island, New York, USA. Emerg. Infect. Dis. 2019;25:792–796. doi: 10.3201/eid2504.181541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heath A. Biology, ecology, and distribution of the tick Haemaphysalis longicornis Nuemann (Acari: Ixodidae) in New Zealand. N. Z. Vet. J. 2016;64:10–20. doi: 10.1080/00480169.2015.1035769. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z., Yang X., Bu F., Yang X., Liu J. Morphological, biological and molecular characteristics of bisexual and parthenogenetic Haemaphysalis longicornis. Vet. Parasitol. 2012;189:344–352. doi: 10.1016/j.vetpar.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 11.Hyeon J.-Y., McGinnis H., Sims M., Helal Z.H., Kim J., Chung D.H., Risatti G.R., Lee D.-H. Complete mitochondrial genome of Asian longhorned tick, Haemaphysalis longicornis, Neumann, 1901 (Acari: Ixodida: Ixodidae) identified in the United States. Mitochondrial DNA B Resour. 2021;6:2402–2405. doi: 10.1080/23802359.2021.1922100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sivakumar T., Hayashida K., Sugimoto C., Yokoyama N. Evolution and genetic diversity of Theileria. Infect. Genet. Evol. 2014;27:250–263. doi: 10.1016/j.meegid.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Jeong W., Yoon S.H., An D.J., Cho S.H., Lee K.K., Kim J.Y. A molecular phylogeny of the benign Theileria parasites based on major piroplasm surface protein (MPSP) gene sequences. Parasitol. 2010;137:241–249. doi: 10.1017/S0031182009991090. [DOI] [PubMed] [Google Scholar]
- 14.Kakuda T., Shiki M., Kubota S., Sugimoto C., Brown W.C., Kosum C., Nopporn S., Onuma M. Phylogeny of benign Theileria species from cattle in Thailand, China and the U.S.A. Int. J. Parasitol. 1998;28:1261–1267. doi: 10.1016/S0020-7519(98)00113-1. [DOI] [PubMed] [Google Scholar]
- 15.Dinkel K.D., Herndon D.R., Noh S.M., Lahmers K.K., Todd S.M., Ueti M.W., Scoles G.A., Mason K.L., Fry L.M. A U.S. isolate of Theileria orientalis, Ikeda genotype, is transmitted to cattle by the invasive Asian longhorned tick, Haemaphysalis longicornis. Parasites Vectors. 2021;14:157. doi: 10.1186/s13071-021-04659-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oakes V.J., Yabsley M.J., Schwartz D., LeRoith T., Bissett C., Broaddus C., Schlater J.L., Todd S.M., Boes K.M., Brookhart M., et al. Theileria orientalis ikeda genotype in cattle, Virginia, USA. Emerg. Infect. Dis. 2019;25:1653–1659. doi: 10.3201/eid2509.190088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson A.T., White S., Shaw D., Egizi A., Lahmers K., Ruder M.G., Yabsley M.J. Theileria orientalis Ikeda in host-seeking Haemaphysalis longicornis in Virginia, U.S.A. Ticks Tick Borne Dis. 2020;11:101450. doi: 10.1016/j.ttbdis.2020.101450. [DOI] [PubMed] [Google Scholar]
- 18.Almazán C., Scimeca R.C., Reichard M.V., Mosqueda J. Babesiosis and Theileriosis in North America. Pathogens. 2022;11:168. doi: 10.3390/pathogens11020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Telionis A., Lahmers K., Todd M., Carbonello A., Broaddus C.C., Bissett C.J., Hungerford L.L. Distribution of Theileria orientalis in Virginia Market Cattle, 2018-2020. Pathogens. 2022;11:1353. doi: 10.3390/pathogens11111353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao L., Li J., Cui X., Jia N., Wei J., Xia L., Wang H., Zhou Y., Wang Q., Liu X., et al. Distribution of Haemaphysalis longicornis and associated pathogens: Analysis of pooled data from a China field survey and global published data. Lancet Planet. Health. 2020;4:e320–e329. doi: 10.1016/S2542-5196(20)30145-5. [DOI] [PubMed] [Google Scholar]
- 21.Panella N.A., Nicholson W.L., Komar N., Burkhalter K.L., Hughes H.R., Theuret D.P., Blocher B.H., Sexton C., Connelly R., Rothfeldt L., et al. Field-Collected Ticks From Benton County, Arkansas, and Prevalence of Associated Pathogens. J. Med. Entomol. 2024;61:1035–1042. doi: 10.1093/jme/tjae040. [DOI] [PubMed] [Google Scholar]
- 22.Price K.J., Ayres B.N., Maes S.E., Witmier B.J., Chapman H.A., Coder B.L., Boyer C.N., Eisen R.J., Nicholson W.L. First detection of human pathogenic variant of Anaplasma phagocytophilum in field-collected Haemaphysalis longicornis, Pennsylvania, USA. Zoonoses Public Health. 2022;69:143–148. doi: 10.1111/zph.12901. [DOI] [PubMed] [Google Scholar]
- 23.Thompson A.T., White S.A., Shaw D., Garrett K.B., Wyckoff S.T., Doub E.E., Ruder M.G., Yabsley M.J. A multi-seasonal study investigating the phenology, host and habitat associations, and pathogens of Haemaphysalis longicornis in Virginia, U.S.A. Ticks Tick Borne Dis. 2021;12:101773. doi: 10.1016/j.ttbdis.2021.101773. [DOI] [PubMed] [Google Scholar]
- 24.Johnson B., Snellgrove A.N., McBride S.E., Hartzer K., Levin M.L., Nicholson W.L. Acquisition of Rickettsia rickettsii (Rickettsiales: Rickettsiaceae) by Haemaphysalis longicornis (Acari: Ixodidae) through co-feeding with infected Dermacentor variabilis (Acari: Ixodidae) in the laboratory. J. Med. Entomol. 2023;60:1380–1387. doi: 10.1093/jme/tjad107. [DOI] [PubMed] [Google Scholar]
- 25.Egizi A.M., Robbins R.G., Beati L., Nava S., vans C.R., Occi J.L., Fonseca D.M. A pictorial key to differentiate the recently detected exotic Haemaphysalis longicornis Neumann, 1901 (Acari, Ixodidae) from native congeners in North America. Zookeys. 2019;818:117–128. doi: 10.3897/zookeys.818.30448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keirans J.E., Litwak T.R. Pictorial key to the adults of hard ticks, family Ixodidae (Ixodida: Ixodoidea), east of the Mississippi River. J. Med. Entomol. 1989;26:435–448. doi: 10.1093/jmedent/26.5.435. [DOI] [PubMed] [Google Scholar]
- 27.Thompson A.T., Dominguez K., Cleveland C.A., Dergousoff S.J., Doi K., Falco R.C., Greay T., Irwin P., Lindsay L.R., Liu J., et al. Molecular Characterization of Haemaphysalis Species and a Molecular Genetic Key for the Identification of Haemaphysalis of North America. Front. Vet Sci. 2020;7:141. doi: 10.3389/fvets.2020.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamau J., de Vos A.J., Playford M., Salim B., Kinyanjui P., Sugimoto C. Emergence of new types of Theileria orientalis in Australian cattle and possible cause of theileriosis outbreaks. Parasit Vectors. 2011;4:22. doi: 10.1186/1756-3305-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamau J., Salim B., Yokoyama N., Kinyanjui P., Sugimoto C. Rapid discrimination and quantification of Theileria orientalis types using ribosomal DNA internal transcribed spacers. Infect Genet. Evol. 2011;11:407–414. doi: 10.1016/j.meegid.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 30.de la Fuente J., Van Den Bussche R.A., Prado T.M., Kocan K.M. Anaplasma marginale msp1alpha genotypes evolved under positive selection pressure but are not markers for geographic isolates. J. Clin. Microbiol. 2003;41:1609–1616. doi: 10.1128/JCM.41.4.1609-1616.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dennis D.T., Nekomoto T.S., Victor J.C., Paul W.S., Piesman J. Reported distribution of Ixodes scapularis and Ixodes pacificus (Acari: Ixodidae) in the United States. J. Med. Entomol. 1998;35:629–638. doi: 10.1093/jmedent/35.5.629. [DOI] [PubMed] [Google Scholar]
- 32.USDA, National Agricultural Statistics Service Cattle Inventory. [(accessed on 19 August 2024)];2023 Available online: https://www.nass.usda.gov/Surveys/Guide_to_NASS_Surveys/Cattle_Inventory/
- 33.Raghavan R.K., Barker S.C., Cobos M.E., Barker D., Teo E.J.M., Foley D.H., Nakao R., Lawrence K., Heath A.C.G., Peterson A.T. Potential Spatial Distribution of the Newly Introduced Long-horned Tick, Haemaphysalis longicornis in North America. Sci. Rep. 2019;9:498. doi: 10.1038/s41598-018-37205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.USDA, National Agricultural Statistics Service State Agriculture Overview: Virginia. [(accessed on 19 August 2024)];2023 Available online: https://www.nass.usda.gov/Quick_Stats/Ag_Overview/stateOverview.php?state=VIRGINIA.
- 35.Lawrence K.E., Forsyth S.F., Vaatstra B.L., McFadden A.M.J., Pulford D.J., Govindaraju K., Pomroy W.E. Clinical Haematology and Biochemistry Profiles of Cattle Naturally Infected with Theileria Orientalis Ikeda Type in New Zealand. N. Z. Vet. J. 2018;66:21–29. doi: 10.1080/00480169.2017.1391142. [DOI] [PubMed] [Google Scholar]
- 36.Neilson F.J.A., Mossman D.H. Anaemia and deaths in red deer (Cervus elaphus) fawns associated with heavy infestations of cattle tick (Haemaphysalis longicornis) N. Z. Vet. J. 1982;30:125–126. doi: 10.1080/00480169.1982.34908. [DOI] [PubMed] [Google Scholar]
- 37.Perera P.K., Gasser R.B., Firestone S.M., Anderson G.A., Malmo J., Davis G., Beggs D.S., Jabbar A. Oriental theileriosis in dairy cows causes a significant milk production loss. Parasites Vectors. 2014;7:73. doi: 10.1186/1756-3305-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McFadden A., Hart M., Bueno I., Ha H., Heath A., Pulford D. Monitoring Theileria orientalis (Ikeda)-associated bovine anaemia in affected cattle over time. Vet. Parasitol. 2017;245:29–33. doi: 10.1016/j.vetpar.2017.07.035. [DOI] [PubMed] [Google Scholar]
- 39.Cumbie A.N., Trimble R.N., Eastwood G. Pathogen Spillover to an Invasive Tick Species: First Detection of Bourbon Virus in Haemaphysalis longicornis in the United States. Pathogens. 2022;11:454. doi: 10.3390/pathogens11040454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poh K.C., Aguilar M., Capelli-Peixoto J., Davis S.K., Ueti M.W. Haemaphysalis longicornis (Acari: Ixodidae) does not transmit Babesia bovis, a causative agent of cattle fever. Ticks Tick Borne Dis. 2024;15:102374. doi: 10.1016/j.ttbdis.2024.102374. [DOI] [PubMed] [Google Scholar]
- 41.Raney W.R., Perry J.B., Hermance M.E. Transovarial transmission of heartland virus by invasive Asian longhorned ticks under laboratory conditions. Emerg. Infect. Dis. 2022;28:726–729. doi: 10.3201/eid2803.210973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otsuka H. Studies on transmission of Babesia gibsoni Patton (1910) by Haemaphysalis longicornis Neuman (1901) Bull. Fac. Agric. Miyazaki Univ. 1974;21:359–367. [Google Scholar]
- 43.Zhuang L., Sun Y., Cui X.M., Tang F., Hu J.G., Wang L.Y., Cui N., Yang Z.D., Huang D.D., Zhang X.A., et al. Transmission of Severe Fever with Thrombocytopenia Syndrome Virus by Haemaphysalis longicornis Ticks, China. Emerg Infect Dis. 2018;24:868–871. doi: 10.3201/eid2405.151435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Butler R.A., Trout Fryxell R.T. Management of Haemaphysalis longicornis (Acari: Ixodidae) on a cow-calf farm in East Tennessee, USA. J. Med. Entomol. 2023;60:1374–1379. doi: 10.1093/jme/tjad121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data from this study are freely available within the GenBank database under accession numbers PQ231312–PQ231315; PQ300672; and PQ301201.