Figure 2.
Proteomic and phosphoproteomic landscape of the post-implantation/pre-gastrulation embryo
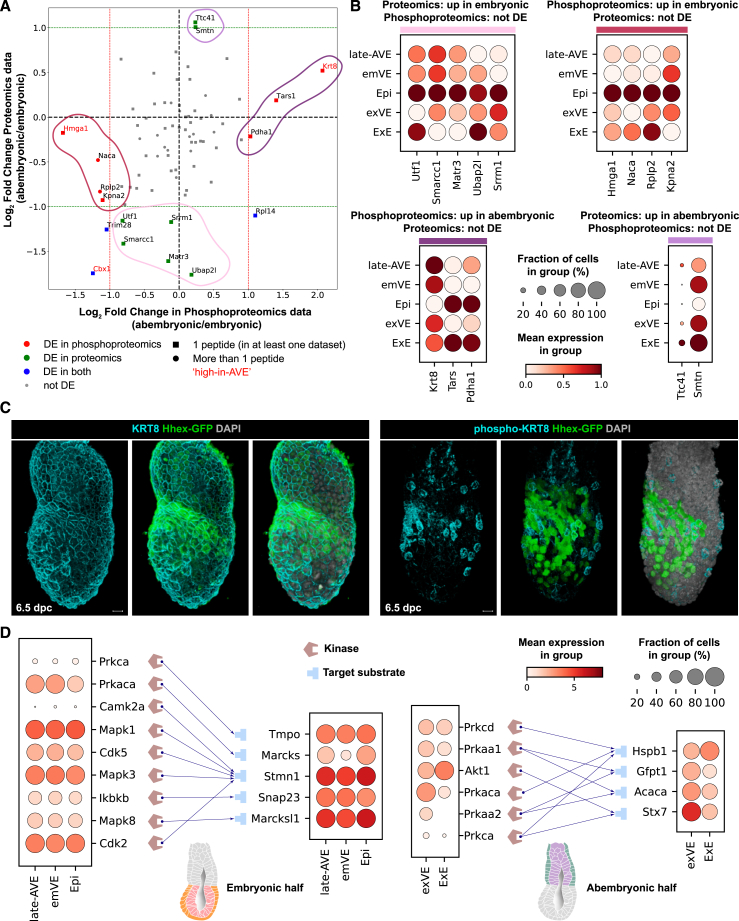
(A) Scatterplot of log2 fold change between the embryonic and abembryonic halves, in proteomics vs. phosphoproteomics data. Each marker represents a protein, and those corresponding to genes from the high-in-AVE group are highlighted in red. Pearson’s correlation coefficient = 0.36; p = 1.3 × 10−3.
(B) Dot plots of gene expression (scRNA-seq) corresponding to (phospho)proteins that are differentially expressed between the embryonic and abembryonic halves only in one dataset (sets outlined in A).
(C) Surface renderings, showing expression of KRT8 (n = 7) and phospho(Ser23)-KRT8 (n = 5) visualized by immunofluorescence in 6.5-dpc embryos. Hhex-GFP marks the AVE. Scale bars represent 20 μm.
(D) Kinase-substrate bipartite networks predicted for substrate proteins upregulated in the embryonic (left) or abembryonic (right) half in the phosphoproteomics dataset. Dot plots of gene expression (scRNA-seq) are shown for the corresponding kinases and substrates.