Figure 4.
Origin and fate of the AVE
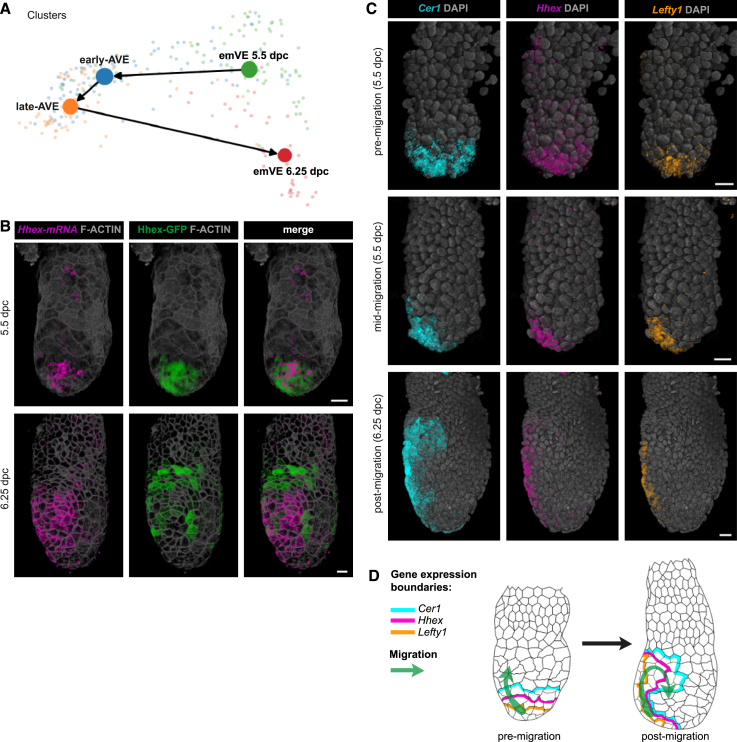
(A) PAGA graph computed from RNA velocities and projected on the first two diffusion components of a diffusion map of the AVE and emVE clusters, combining the data from both stages.
(B) Direct comparison of short-term lineage-labeled AVE cells (expressing the Hhex-GFP reporter) and the contemporaneous expression of the endogenous Hhex transcript (visualized by HCR) in 5.5-dpc (n = 7) and 6.25-dpc (n = 5) embryos. Embryos are rotated by ∼20° about their PD axis to show the full anterior surface along with lateral sides.
(C) Volume renderings directly comparing changes to the expression domains of the AVE markers Cer1, Hhex, and Lefty1 in embryos before (n = 13), during (n = 18), and after (n = 8) AVE migration. Embryos are orientated anterior to the left.
(D) Schematic summarizing changes to Cer1, Hhex, and Lefty1 expression domains throughout the VE because of AVE migration. Scale bars represent 20 μm.