Figure 5.
Spatial mapping of the transcriptional heterogeneity seen within the AVE and exVE clusters
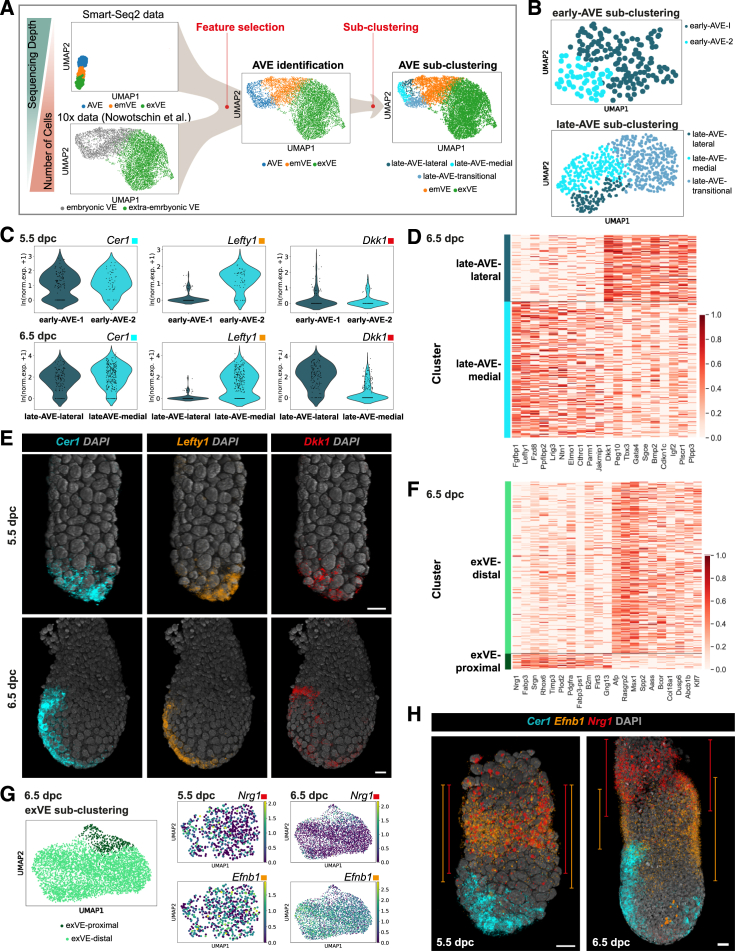
(A) Computational strategy used for VE sub-clustering of 10× scRNA-seq data (Nowotschin et al.17), using gene expression features extracted from our Smart-seq2 data. The AVE cluster was selected for further sub-clustering analysis.
(B) UMAP plots of cells (data from Nowotschin et al.17) belonging to the AVE at 5.5 dpc (top) and 6.5 dpc (bottom), colored according to sub-clusters (Figure S4 and STAR Methods for AVE identification in these data).
(C) Violin plots showing Cer1, Lefty1, and Dkk1 normalized log expression in cells grouped according to the clusters in (B), for both stages.
(D) Heatmap showing normalized log expression of the top genes upregulated in the late-AVE-medial cluster and the AVE-lateral cluster (10 each), obtained through a differential expression analysis between the two clusters at 6.5 dpc.
(E) Volume renderings showing the expression domains of Cer1, Lefty1, and Dkk1, used to distinguish between the sub-clusters of the AVE at 5.5 (n = 8) and 6.5 (n = 3) dpc.
(F) Heatmap showing normalized log expression of the top 10 genes upregulated in the exVE-proximal cluster and the exVE-distal cluster at 6.5 dpc. Nrg1 (which ranked 13th in the exVE-proximal cluster by adjusted p value) was manually added to the list.
(G) UMAP plots showing sub-clusters within the exVE at 6.5 dpc (left) and of cells belonging to the exVE cluster at 5.5 (center) and 6.5 dpc (right), colored according to normalized log expression of Nrg1 and Efnb1.
(H) Volume renderings showing changes to the expression domains of Efnb1 and Nrg1 between 5.5 (n = 6) and 6.5 (n = 4) dpc in comparison with Cer1-expressing AVE cells. Orange and red lines represent, for Efnb1 and Nrg1, respectively, the proximal-to-distal extent of their expression domains. Scale bars represent 20 μm, and embryos are orientated anterior to the left.