Figure 6.
Semaphorin signaling is required for correct AVE migration
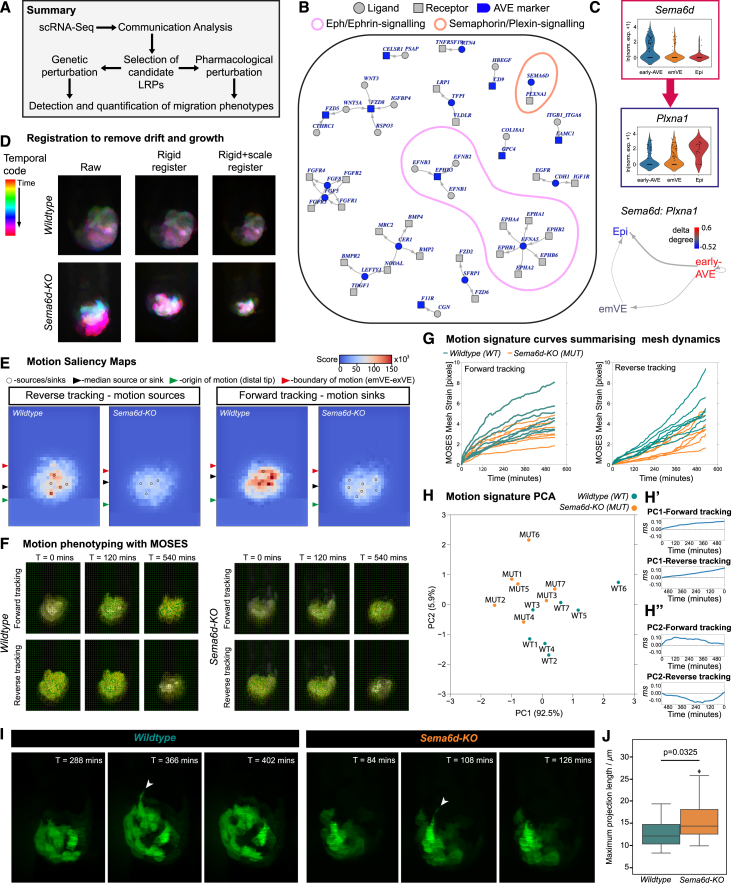
(A) Schematic of computational and experimental strategy used to discover signaling pathways important for AVE migration.
(B) Plot showing ligand-receptor pairs (LRPs) containing at least one high-in-AVE component (blue), at 5.5 and/or 6.25 dpc. Arrows indicate the direction of signaling.
(C) Violin plots showing normalized log expression levels of Sema6d and Plxna1 in AVE, emVE, and Epi clusters, at 5.5 dpc (top), and the intercellular communication pattern associated with SEMA6D:PLXNA1 (bottom).
(D) Rigid body and scaling registration used to remove drift and growth from confocal time-lapse movies for phenotyping of AVE migration with MOSES analysis.
(E) MOSES motion saliency maps computed from reverse and forward tracking (STAR Methods) to identify spatial location of motion sources (left) and sinks (right) from a wild-type and Sema6d homozygous mutant (Sema6d-KO) embryo. The position of the median source/sink relative to the origin and boundary of motion is marked.
(F) Snapshots of the mesh connecting initially neighboring superpixels when MOSES is applied to track AVE migration (forward and reverse) in a wild-type and Sema6d-KO embryo.
(G) Plots of motion signatures (mesh strain vs. time) extracted from tracking AVE migration in wild-type and Sema6d-KO embryos (n = 7 each), which summarize the extent of mesh deformation based on forward and reverse tracking.
(H) Plot of the first two principal components (PC1 vs. PC2) after applying principal-component analysis to the values of the mesh strains extracted from the concatenated forward and reverse time-lapse tracking of AVE migration in wild-type (WT) and Sema6d-KO (MUT) embryos. In parentheses is the percentage of explained variance. (H′) and (H″) represent the loadings of PC1 and PC2, respectively, for the mesh strains obtained from the forward (top) and the reverse (bottom) tracking. The monotonic increase of PC1 suggests that it represents overall AVE movement. The non-monotonic behavior of PC2, with a stationary point at ∼120 min, indicates that it represents the rate at which AVE cells slow down.
(I) Maximum intensity projections of high-resolution lattice light-sheet time-lapse frames from cultured wild-type and Sema6d-KO embryos, showing the emergence and retraction of basal projections by migrating Hhex-GFP-positive AVE cells (green). Arrowheads point to basal projections at their measured maximum lengths.
(J) Plot showing difference (p = 0.03; nested ANOVA) in the maximum length of basal projections from wild-type (n = 9 embryos, n = 76 projections) and Sema6d-KO mutant embryos (n = 9, n = 66 projections). Data are represented as mean ± SEM.