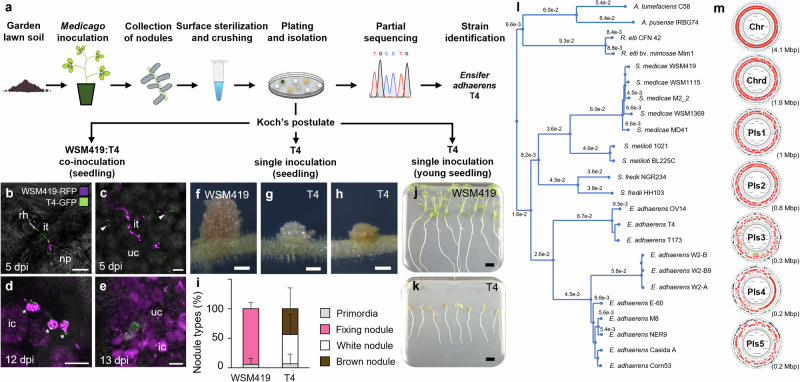
Fig. 1. T4 is an Ensifer adhaerens strain co-colonizing nodules with effective rhizobia, inducing ineffective nodules by itself and triggering the death of its host.
a Schematic procedure used to trap Medicago littoralis R108 (R108) nodule endophytes and to validate the Koch’s postulate. b–e When Ensifer adhaerens T4-GFP (T4-GFP, green signal) is co-inoculated with the efficient symbionts Sinorhizobium medicae WSM419-RFP (WSM419-RFP, violet signal), T4 behave as a nodule endophyte and co-colonized M. truncatula A17 (A17) nodules. b A 5-dpi root hair showing both WSM419-RFP and T4-GFP in a single infection thread. c WSM419-RFP and T4-GFP in common infection threads (white arrowheads), within a 5-dpi nodule primordium. d 12-dpi nodule cells showing the release of WSM419-RFP and T4-GFP bacteroids from infection threads in common infected-cells (white asterisks). e 13-dpi nodule infected cells containing both WSM419-RFP and T4-GFP. (Scale bars b, c, e, 20 µm; d, 50 µm). rh, root hair; it, infection thread; np, nodule primordium; uc, uninfected cell; ic, infected cell; dpi, days post-inoculation. For b-e, similar results were observed in three independent experiments. f-i T4 induced, by itself, the formation of nodules on A17 seedlings. f 14-dpi WSM419 nodule. g, h 14-dpi T4 nodules. (Scale bar f–h, 500 µm). For f–h, similar results were observed in four independent experiments. i Percentage of nodule types present on WSM419- and T4-inoculated plants shown as mean percentage ± s.d. (n = 24 plants). j and k T4 triggered the death of A17 young seedlings. WSM419- (j) and T4- (k) infected plants at 21 dpi. (Scale bars j and k, 1 cm). l Neighbor-joining phylogenetic tree based on whole bacterial genomes including that of T4 and of other strains of the same species, genus or order. m Organization of the T4 genome. The T4 genome is composed of seven circular replicons, one chromosome (Chr), one chromid (Chrd), and five plasmids (Pls1 to Pls5). Plasmid sizes are indicated. A remarkable feature of the T4 genome is the heterogeneous density of repeated regions along the different replicons. Especially, Pls3 harbors 27% of repeated sequences, mostly transposase encoding genes, whereas repeated sequences represent less than 3% of the genome in the other replicons (Supplementary Fig. 2; Supplementary Fig. 3). Source data for Fig. 1i are provided in Source Data file.