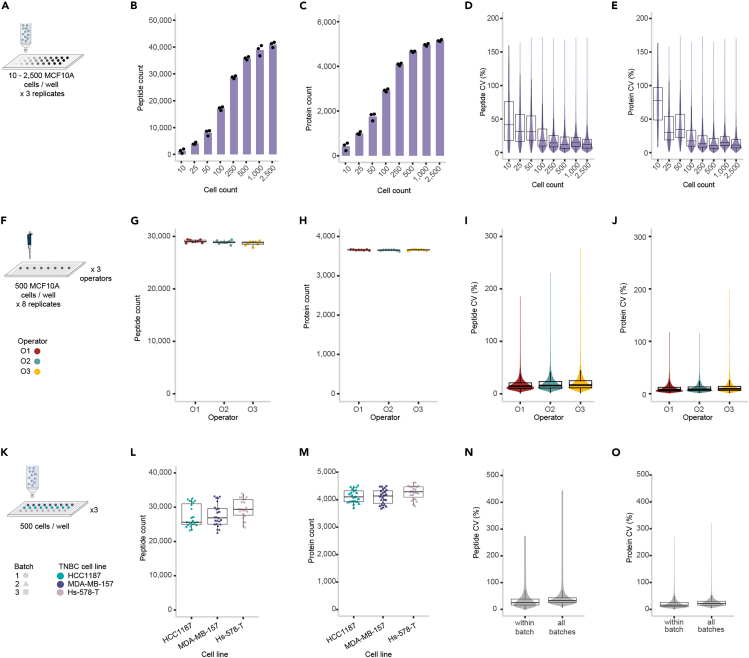
Figure 2.
Expected results for implementing DROPPS on mammary epithelial cell lines
(A) Cell number titration workflow where 10–2,500 MCF10A cells were deposited per well (n = 3 each) by fluorescent-activated cell sorting and subsequently processed using DROPPS.
(B and C) The (B) peptide counts and (C) protein counts of the individual runs where columns represent the mean values.
(D and E) Distribution of intensity coefficients of variation (CVs) at the (D) peptide and (E) protein level.
(F) Schematic of operator experimental design where 500 MCF10A cells were deposited per well (n = 8 each) by three operators and subsequently processed using DROPPS.
(G and H) The (G) peptide counts and (H) protein counts of the individual samples prepared by each operator.
(I and J) Distribution of (I) peptide intensity or (J) protein intensity CVs.
(K) Workflow for testing inter-batch variation by DROPPS. Sorted cells (500 cells) from three TNBC cell lines were processed separately on different days. Each cell line had seven or eight sample replicates processed each day.
(L and M) The (L) peptide counts and (M) protein counts of the individual samples prepared by each operator.
(N and O) Distribution of (N) peptide intensity or (O) protein intensity CVs within a batch or for combining batches. Boxplots show the median, interquartile ranges, and 95% confidence interval estimate. TNBC: triple negative breast cancer. Altered content from Figures S1 and S2 from Waas et al.1