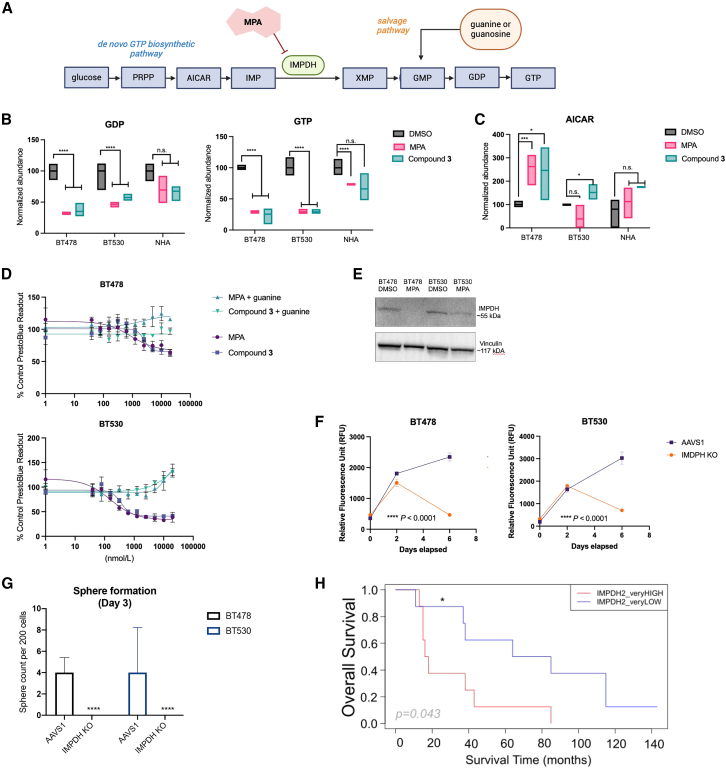
Figure 5.
MPA targets the de novo GTP synthesis pathway in BMICs
(A) Schematic of the de novo GTP synthesis pathway. Created with BioRender.com.
(B) Boxplots depicting relative GDP and GTP levels in patient-derived BMIC lines and normal human astrocytes (NHAs) with either DMSO, MPA, or Compound 3 treatment. Comparisons were made via a two-tailed unpaired t test, n = 4 technical replicates, p value is indicated. N.s., not significant, ∗∗∗∗ = p < 0.0001.
(C) Boxplots depicting relative AICAR levels in patient-derived BMIC lines and NHAs with either DMSO, MPA, or Compound 3 treatment. Comparisons were made via a two-tailed unpaired t test, n = 4 technical replicates. N.s., not significant.
(D) Dose-response curves of BT478 and BT530 tumor cells treated with MPA or Compound 3 in culture media supplemented with exogenous guanine (12 μM) or vehicle (water). Data are presented as mean ± SD from 4 technical replicates.
(E) Immunoblot confirmation of IMPDH (IMPDH1 + IMPDH2) knockout (KO) in patient-derived lung-BMICs. Cropped from single full blot.
(F) Time-course proliferation assays of AAVS1 and IMPDH KO BT478 and BT530 cells. Comparisons were made via a two-tailed unpaired t test. Data are presented as mean ± SD from 4 technical replicates, ∗∗∗∗ = p < 0.0001.
(G) Representative bar graphs depicting the sphere count per 200 cells of AAVS1 vs. IMPDH KO cells. Comparisons were made via a two-tailed unpaired t test, data are presented as mean ± SD from 4 technical replicates, ∗∗∗∗ = p < 0.0001.
(H) Kaplan-Meier curves showing the overall survival probability (from primary cancer diagnosis to death) in a cohort of 30 patients with lung carcinoma who developed BM, for IMPDH2 very high vs. very low expression (1st and 3rd quartiles cutoff values: <114.86 and >210.74). The Cox regression model for survival analysis was used, p values (p) are shown. The hazard ratio (HR) and 95% confidence interval (CI) are HR = 0.30, CI (0.09–0.96) for IMPDH2 very low compared with very high, or inverted HR = 3.33, CI (1.041–11.11). For the two patients from the cohort who were still alive at the end of the study, the latest survival time point was used for the analysis. Data were obtained from the processed (Q3 method normalization) GEO dataset (GSE200563). Data acquisition, analysis, and visualization performed using R version 4.1.2 and the following packages: GEOquery, survival, and ggplot2.