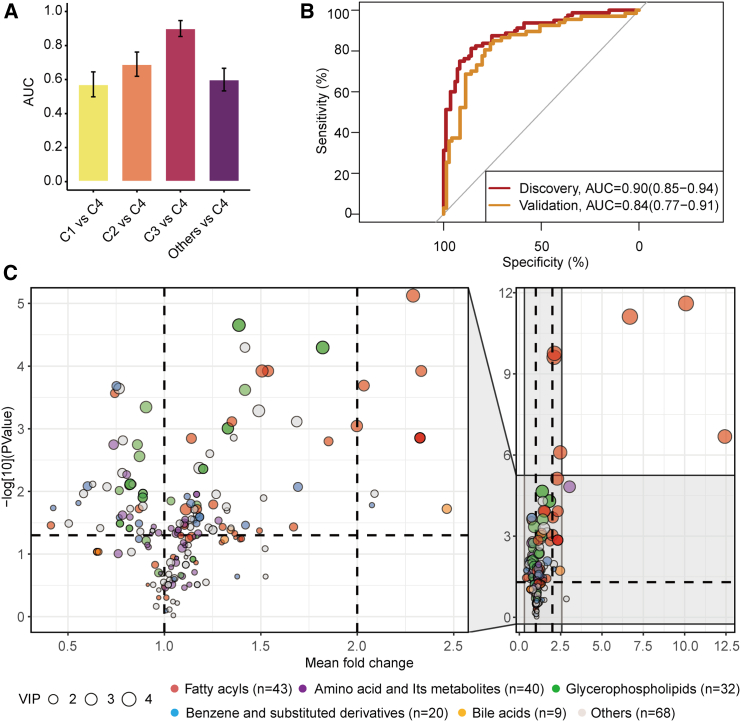
Figure 5.
Association of the gut mycobiome with serum metabolites
(A) Comparing the performance of metabolome-based classifiers (LightGBM) in categorizing participants’ fungal clusters at the baseline (n = 911). Performance of the classifiers was assessed by area under the receiver operating characteristic curve (AUC) across 10-folds, with cluster 4 as the reference group, and each of other groups and their combinations as the comparison group. Here, the numbers of participants included in the cluster 1 to 4 were 518, 227, 85, and 80, respectively.
(B) Receiver operating characteristic curve for the orthogonal projections to latent structures discriminant analysis (OPLS-DA)-selected metabolites in categorizing participants into either cluster 3 or cluster 4 groups within the discovery (n = 165) and validation cohorts (n = 138). The effectiveness of these selected metabolites in predicting fungal clusters was assessed using the machine learning model (LightGBM).
(C) Fold change of differential metabolites between the fungal cluster 3 and cluster 4 (n = 165). The identified metabolites are visualized as dots, with colors corresponding to their respective categories. The dot size indicates the importance of the metabolite in distinguishing fungal clusters during the OPLS-DA analysis.