Figure 1.
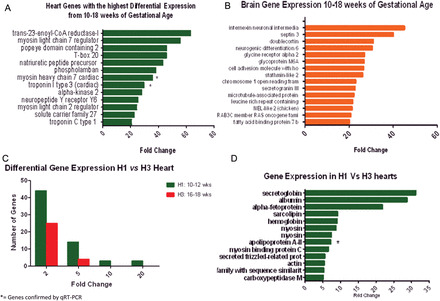
(A) Genes with the highest differential expression in the heart. This figure is based on pooled tissue samples from 14 fetal hearts from 10 to 18 weeks of gestational age for genes with 20-fold change or greater expression level. Human Gene 1.0 ST array (Affymetrix) was used. Genes were chosen based on fold change compared with mRNA, after confirming a differential expression >95% with an false discover rate (FDR) of 5%. * = genes verified by qRT–PCR. (B) Genes with the highest differential expression in the brain. This figure is based on pooled tissue samples from 10 fetal brains from 10 to 18 weeks of gestational age for genes with 20-fold change or greater expression level. Human Gene 1.0 ST array (Affymetrix) was used. Genes were chosen based on fold change compared with mRNA, after confirming a differential expression >95% with an FDR of 5%. * = genes verified by qRT–PCR. (C) Differentially expressed genes measured by fold change in mRNA levels between the hearts at different gestational ages. The late first trimester hearts (H1) showed an increased number of differentially expressed genes (green bars) when compared with the early second trimester hearts (H3). Tissue samples correspond to hearts from 10–12 weeks of gestational age (H1) and 16–18 weeks of gestational age (H3). Differences are measured by fold change (>2), after adjusting for an FDR of 5% with a differential expression >95% using EBarrays (Newton et al., 2001; Kendziorski et al., 2003), to identify differentially expressed genes in any two conditions. (D) Genes with the highest differential expression in the late first trimester hearts (H1). Tissue samples correspond to hearts from 10–12 weeks of gestational age (H1) and 16–18 weeks of gestational age (H3) pooled on two separate Human Gene 1.0 ST array, according to gestational age. Differences are measured by fold change (>2), after adjusting for an FDR of 5% with a differential expression >95% using EBarrays (Newton et al., 2001; Kendziorski et al., 2003), to identify differentially expressed genes in any two conditions. * = gene verified by qRT–PCR.
