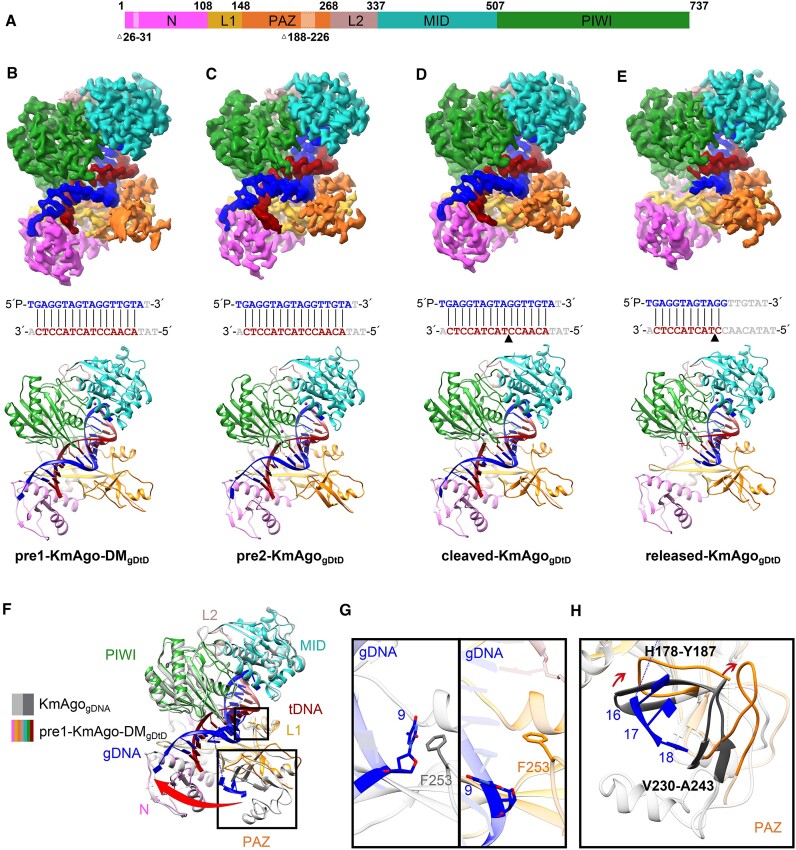
Figure 3.
The overall structures of KmAgo-DMgDtD and KmAgogDtD. (A) Domain architecture of ternary complexes with amino acid numbering. Domains and their unstructured residues are colored and labeled. (B–E) Cryo-EM density map (Top) and ribbon diagrams (Bottom) of pre1-KmAgo-DMgDtD (B), pre2-KmAgogDtD (C), cleaved-KmAgogDtD (D), released-KmAgogDtD (E). (Middle) The sequence and pairing of gDNA (blue) and tDNA (red) strands in the ternary complex. Nucleotides that were not modelled are represented in gray. The black triangle in (D) and (E) denotes the location of the cleavage site. (F) Superposition of pre1-KmAgo-DMgDtD (colored as in B) and KmAgogDNA (gray). (G) Enlarged view of the interaction between position 9 and F253. The left panel is the binary structure, and the right panel is the pre1-KmAgo-DMgDtD. (H) Significant conformational changes in the PAZ domain are indicated by red arrows.