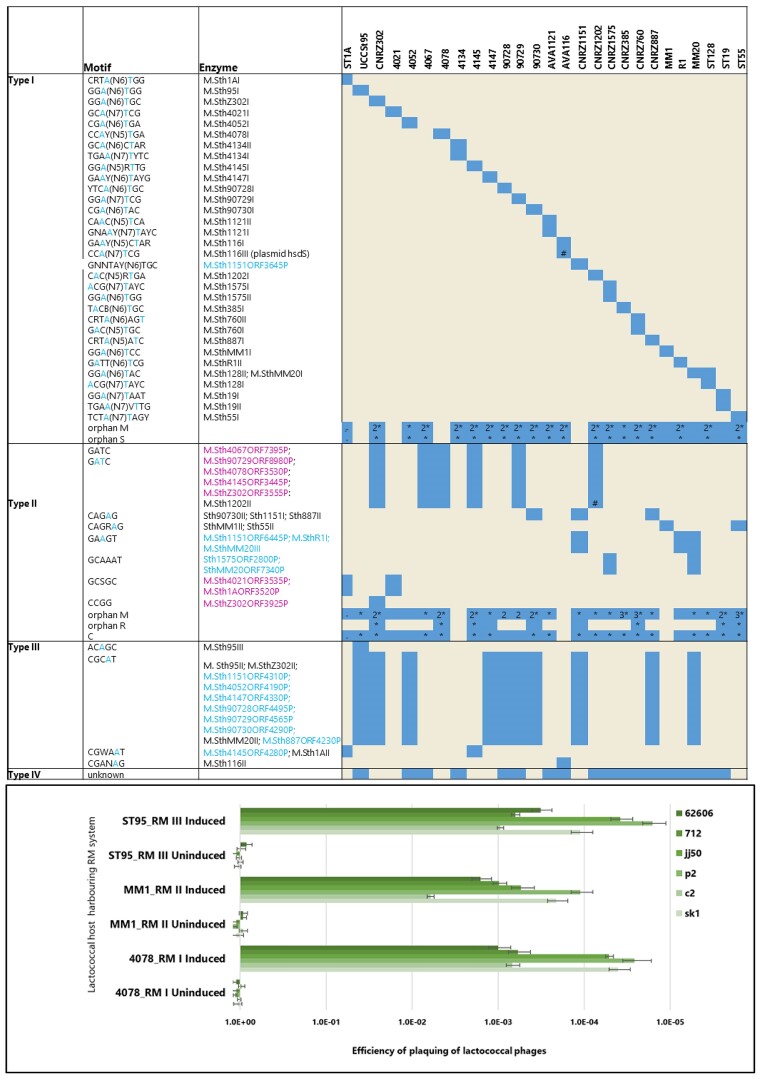
Figure 2.
Upper panel: Identified R/M systems (both active systems and inactive systems/individual genes). Methylated bases within the detected motifs based on SMRT sequencing are indicated in blue text in the ‘motif’ column. Predicted m5C (pink text) and m6A (blue text) associated enzymes which were predicted but where methylation activity was not detected in this study. Where more than one gene was identified, the number of such genes is indicated by a number in the relevant box. In the ‘Enzyme’ column, black text indicates enzymes for which associated methylated motifs were identified, blue and pink text indicates enzymes for which m6A and m5C recognition sequences, respectively, were predicted but methylated motifs were not detected. Lower panel: Efficiency of plaquing (EOP) of lactococcal phages in the presence (induced) or absence (uninduced) of Type I, Type II or Type III R/M systems. In the uninduced state, there is no observable phage-resistance while in the induced state, all three evaluated R/M systems were observed to elicit substantial resistance against the evaluated phages 62606, 712, jj50, p2, c2 and sk1.