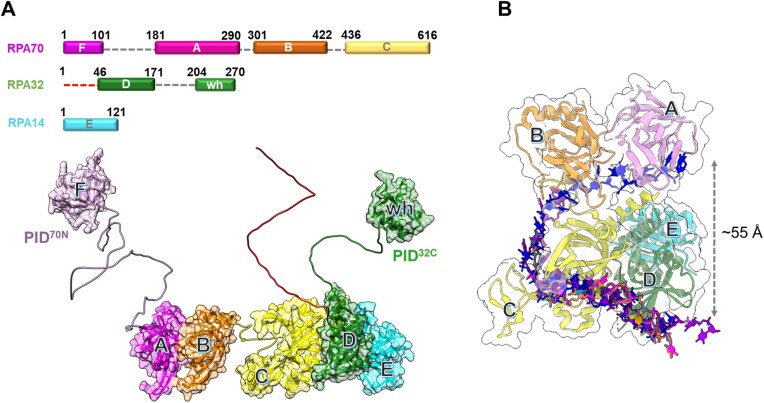
Figure 1.
Architecture of the replication protein A (RPA) – ssDNA complex. (A) The three subunits of RPA—RPA70, RPA32 and RPA14 house several oligonucleotide/oligosaccharide binding (OB) domains that are classified as either DNA binding domains (DBDs: DBD-A, DBD-B, DBD-C and DBD-D) or protein-interaction domains (OB-F/PID70N and winged helix (wh)/PID32C). A structural model of RPA is shown and was generated using information from known structures of the OB-domains and AlphaFold2 models of the linkers. (B) Crystal structure of Ustilago maydis RPA bound to ssDNA is shown (PDB 4GNX). This structure lacks OB-F, the F-A linker, wh and the D-wh linker. ssDNA from CryoEM structures of Saccharomyces cerevisiae RPA (PDB 6I52) and Pyrococcus abyssi RPA (8OEL) are superimposed and reveal an end-to-end DNA distance of ∼55 Å. Structural data supports partial wrapping of ssDNA around the OB domains.