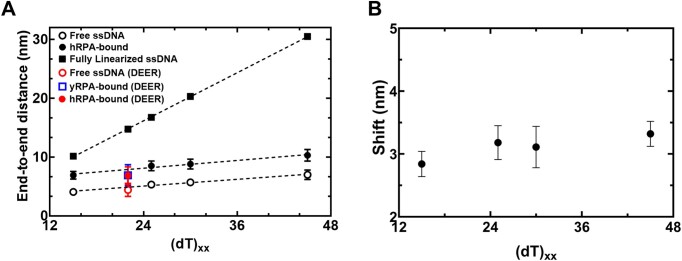
Figure 4.
RPA binding produces a modest 3 nm increase in end-to-end distance. (A) The estimated end-to-end distance between the 3' and 5' ends of ssDNA is plotted as a function of nucleotides which make up the chain. Closed squares represent theoretically calculated end-to-end distances assuming the ssDNA was completely linearized. Open and closed circles represent experimental end-to end distance measurements from smFRET analysis ssDNA -/+ RPA, respectively. The figure also shows that end-to-end distances of yRPA-bound, hRPA-bound, and free (dT)22 ssDNA, measured using DEER spectroscopy. The DEER measurements fall on the respective trend lines suggesting good agreement between experimental measurements performed using two independent biophysical approaches. (B) A ∼3.1 ± 0.2 nm shift is observed in the end-to-end distances between the free ssDNA and RPA-bound ssDNA. This ssDNA-independent uniform expansion suggests that the average dimensions of the DNA ensemble are increasing due to interactions with RPA.