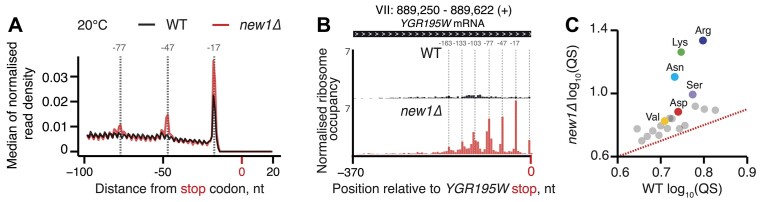
Figure 3.
New1 loss is associated with ribosomal queuing upstream of stop codons proceeded by C-terminal arginine, lysine and asparagine. (A) Metagene analysis of the 5′ read ends for the 5PSeq libraries generated from wild-type (WT MYJ1171, black trace) and New1-deficient (new1Δ MYJ1173, red trace) yeast strains grown at 20°C. Stalls represent the 5′ boundary of the ribosome-protected 5′-phosphorylated mRNA fragment. The 5′ position is offset by -17 nt from the first nucleotide in the ribosomal A site. With an 80S ribosome covering 30 mRNA bases, the 30-nt periodicity of the in the new1Δ metagene is indicative of multiple ribosomes queued at the stop codon. (B) An example of a heavily ‘queued’ ORF YGR195W encoding a C-terminal AGG arginine codon. Normalised read coverage around the stop codon is shown. (C) Ribosomal queuing scores (QS) calculated for ORFs parsed by the codon preceding the stop codon. The red dashed line is a guide for the expected position of data points in the absence of the systematic difference between in QS between the wild-type and new1Δ samples. All analyses were performed on pooled wild-type and new1Δ 5PSeq datasets from three biological replicates each.