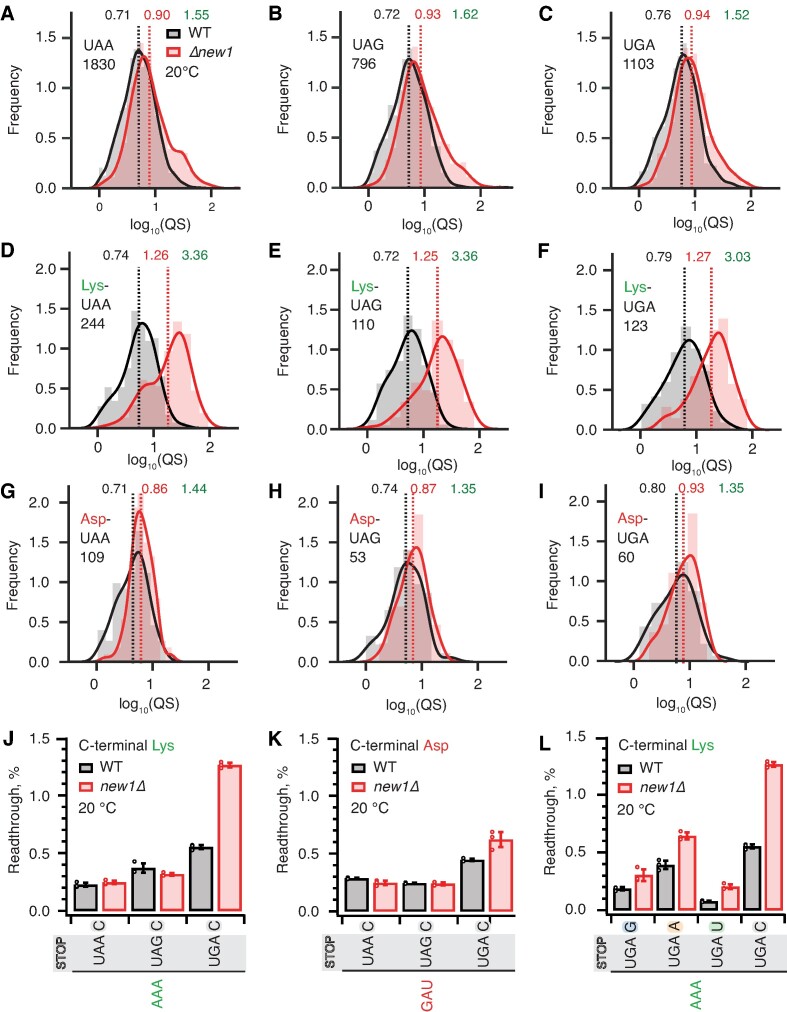
Figure 4.
The absence of New1 specifically increases ribosomal readthrough when the weak stop codon UGA is preceded by C-terminal lysine. (A–C) Binned distributions of ribosome queuing scores (QS) for ORFs terminating in either UAA (A), UAG (B) or UGA (C) stop codons. (D–F) Same as (A–C), but with for ORFs encoding C-terminal lysine residues. Geomeans of QS distributions for wild type is black and for new1Δ is in red, the QS fold change is in green. Numbers of ORFs included in the analyses are shown on the figure, e.g. 1830 instances of ORFs with UAA stop codon. All analyses were performed on pooled datasets from three replicates collected at 20°C. (G, H) Same as (D-F), but with for ORFs encoding C-terminal aspartic acid. (J, K) Stop codon readthrough efficiencies in wild-type and new1Δ strains measured with dual-luciferase reporters harbouring UAA, UAG and UGA stop codons in combination with either a C-terminal AAA lysine codon (J) or GAU aspartic acid codon (K). (L) Readthrough efficiency of UAG G, UAG A, UAG U and UAG C extended stop codons in combination with a C-terminal AAA lysine codon in both wild-type and new1Δ strains. Note that the same AAA UGA C results are shown on both panels (J) and (L) in order to make the data comparison easier. Error bars represent standard deviations. All experiments were performed at 20°C.