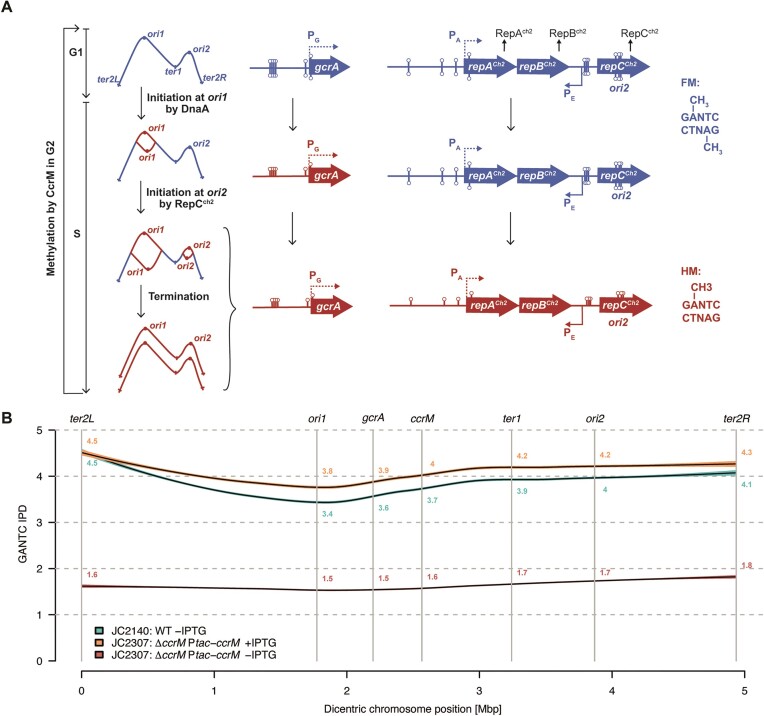
Figure 1.
Methylation of GANTC motifs on the dicentric chromosome of A. tumefaciens. (A) Schematic showing the predicted methylation state of GANTC motifs depending on their chromosomal location and on cell cycle progression. ‘FM’ stands for fully-methylated DNA (blue color) with m6A on both GANTC strands and ‘HM’ stands for hemi-methylated DNA (red color) with m6A on only one GANTC strand (the newly-replicated strand is not yet methylated by the CcrM MTase). The right side of this schematic focuses on the predicted methylation state of the gcrA and repABCCh2 loci as a function of cell cycle progression, which depends on their chromosomal position. Lollipops indicate m6A in GANTC motifs (including 200 bp upstream of ORFs). (B) CcrM-dependent methylation of GANTC motifs on the A. tumefaciens dicentric chromosome. gDNA samples from WT and JC2307 (ΔccrM Ptac-ccrM) cells that grew exponentially in ATGN ± IPTG for 7 h were analyzed by SMRT-seq. GANTC IPD ratios were directly generated by the PacBio software. A smooth curve was then fitted between these ratios and the dicentric chromosome position using the LOESS function of R with default parameters. The 95% confidence interval was then plotted to predict average GANTC IPD ratios depending on their position on the dicentric chromosome from ter2L (position ‘0 Mbp’) to ter2R (position ‘4.936 Mbp’).