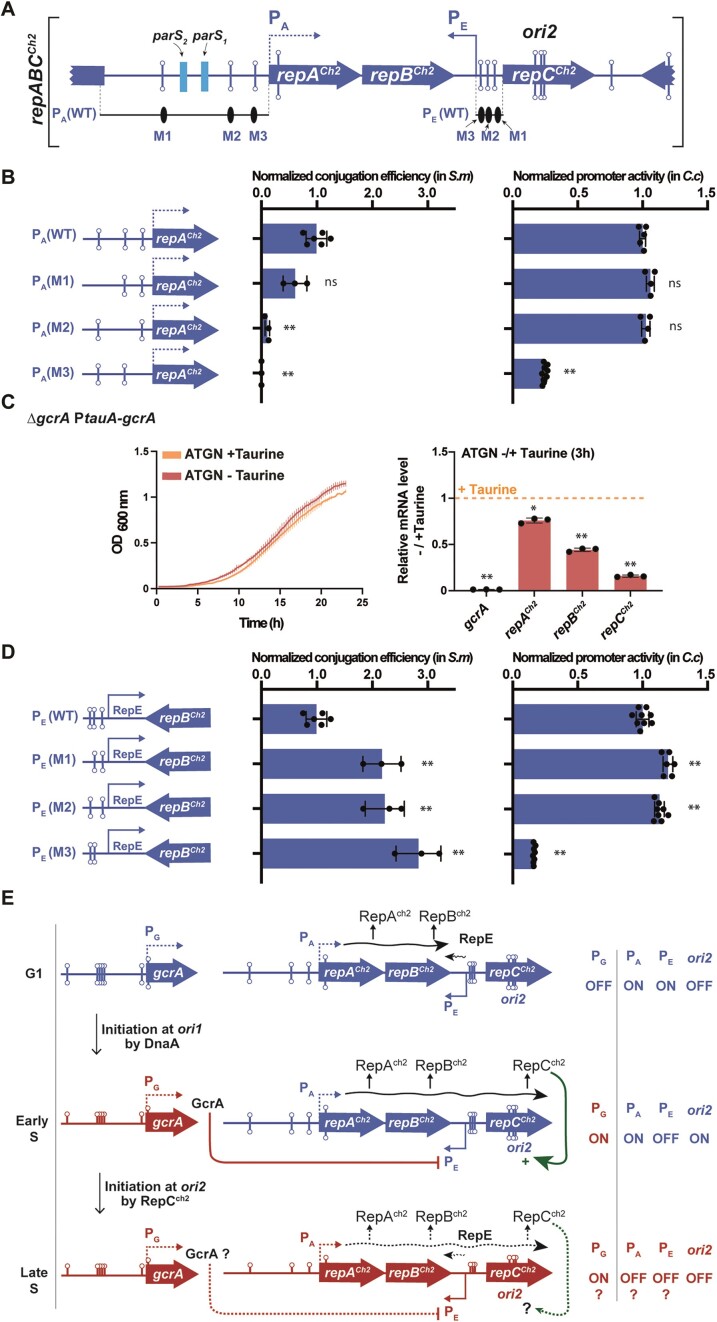
Figure 6.
The RepABCCh2/ori2 module is sufficient for autonomous replication and the expression of its RepCCh2 initiator is activated by GcrA. (A) Schematic of the repABCCh2/ori2 module showing where ORFs (blue arrows), GANTC motifs (lollipops) and predicted parS sites (light blue rectangles) (9) are located. Note that the three ORFs are not drawn to scale to better visualize the organization of IG regions. This schematic shows the exact WT module cloned into pSW25T for the conjugation assays shown in the left panels of (B) and (D). It also shows the PA(WT) and PE(WT) promoter regions cloned into pOT1e for the promoter activity assays shown in the right panels of (b) and (d), respectively. (B) Impact of the three GANTC motifs found in the PA(WT) promoter on plasmid replication/maintenance in S. meliloti cells (left) and on promoter activity in C. crescentus cells (right). For the PA(M1), PA(M2) and PA(M3) derivatives of PA(WT), single methylatable GANTC motifs were mutated into non-methylatable GTNTC motifs. Means from minimum 3 independent experiments were plotted for each construct. (C) The impact of GcrA on repABCCh2 expression in A. tumefaciens. Left panel: JC2899 (ΔgcrA PtauA-gcrA) cells were pre-cultured into ATGN + Taurine. Cells were then washed and diluted to an OD600∼0.05 into ATGN ± Taurine and placed into a plate reader to record OD600 every 15 min. Error bars represent means ± standard deviations from three biologically independent replicates. Right panel: JC2899 cells were pre-cultured in ATGN + Taurine. Once cells reached exponential phase, they were then washed and resuspended to an OD600∼0.2 into ATGN ± Taurine. RNA samples were prepared after 3h of growth for qRT-PCR analyses. The graph shows relative mRNA levels in cells cultivated without the Taurine inducer, compared to cells cultivated with the Taurine inducer (then set to an arbitrary value of 1 for each gene = orange dotted line). Three biological replicates with three technical replicates were used for each condition. Significant differences (Wilcoxon rank sum test) when comparing -Taurine/+Taurine are indicated by * (P< 0.05) and ** (P< 0.01). (D) Impact of the three GANTC motifs found in the PE (WT) promoter on plasmid replication/maintenance in S. meliloti cells (left) and on promoter activity in C. crescentus cells (right). For the PE(M1), PE(M2) and PE(M3) derivatives of PE(WT), single methylatable GANTC motifs were mutated into non-methylatable GTNTC motifs. For (B) and (D), minimum 3 independent samples (dots) were used for each construct. Conjugation efficiencies and promoter activities were normalized so that the mean value of the WT construct equals one in each case. Mean values were plotted with error bars corresponding to standard deviations. Student's t-tests comparing mutant constructs with WT constructs: ns = not significant (P-value > 0.05), ** = P-value < 0.001.(E) Model for the cell cycle-dependent regulation of the repABCCh2/ori2 module by GcrA. Dashed arrows or ‘?’ indicate still hypothetical regulatory pathways. Wavy lines indicate when transcription from PA (promoter region upstream of the repABCCh2operon) or PE (promoter driving RepECh2 expression) is expected to take place based on their methylation states. When the TSS could not be predicted (for PA and PG), it was arbitrarily positioned at the beginning of the ORF.