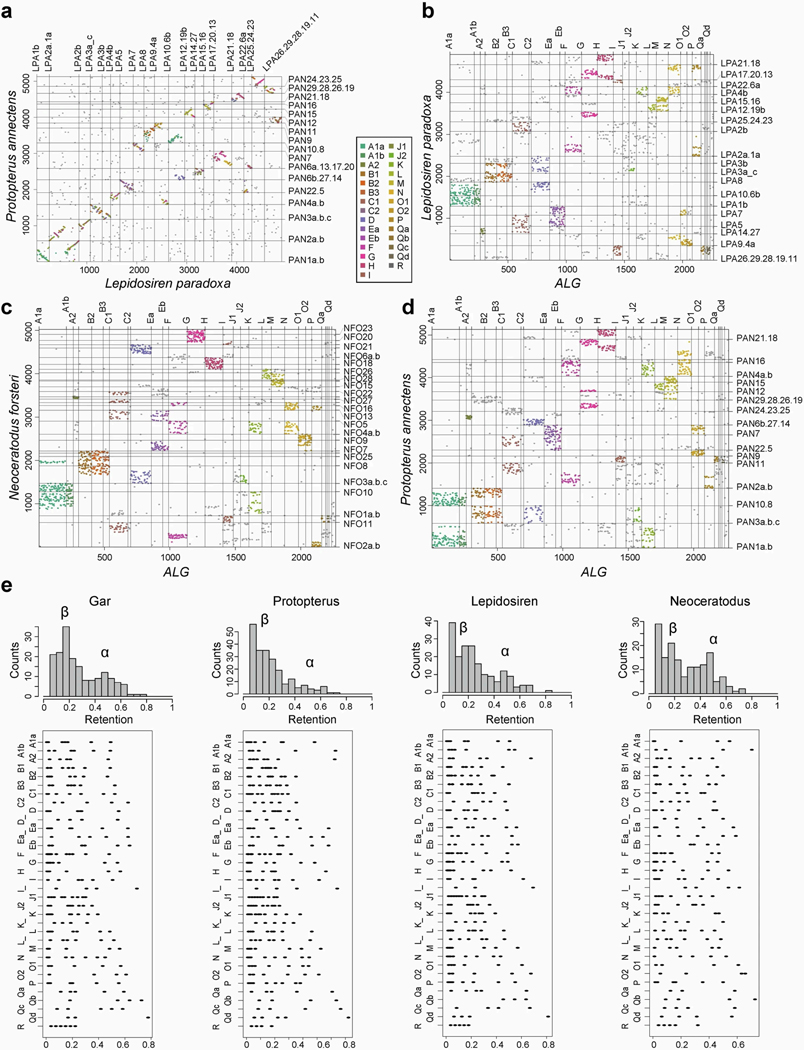
Extended Data Fig. 2: High retention of ancestral linkage groups lungfish genomes.
a-d, Species-to-species dotplots showing high degree of retained collinearity in the African and South American lungfish genomes, despite their genome size. b-d, Oxford dotplots representing orthologous genes shared on the previously reported ancestral linkage groups (ALGs)15. Chromosome numbering corresponds to the homologous lungfish linkage groups which have independently fused in individual lineages. Neoceratodus with its 27 chromosomes represented the most ancestral (unfused) state. e, Retention rates of lungfish chromosomes. Often only one alpha copy is present in lungfishes, e.g. descendants of several chromosomal elements have two alpha chromosomes in gar and Australian lungfish but only one clear alpha chromosome remains in South American and African lungfish (with the alpha copies having lost genes). Retention rates were computed as the percentage of the retained (present) ohnologs of gene families that comprise a given ancestral linkage group. Total number of gene families per chromosome was counted and their position was not taken into account. Only chromosomes with at least 5% ancestral linkage group retention were counted. Lower plots show retention on individual chromosomes (represented by dots) grouped by their ancestral linkage group in different lungfishes and gar.