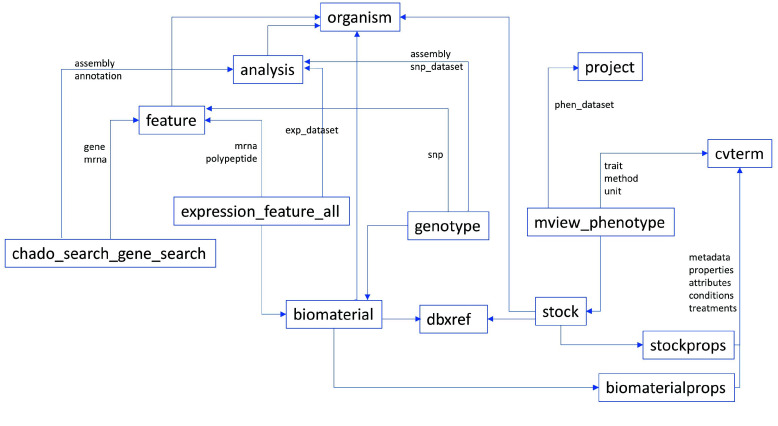
Figure 3.
Entity relationship diagram of Chado tables and materialized views involved in omics integration.
The Tripal modules generate materialized views to aggregate data for genomic features and gene annotations, expression levels, and phenotype values. Genotype metadata on biomaterials and features are also in the database, while the allele values are in external vcf.gz files. Biomaterial and Stock are synchronized through dbxref using NCBI SRR or SAMN accessions when available.